High-sensitivity amplicon library building kit, library building method and application
A kit and amplicon technology, which is applied in the field of sequencing library construction, can solve the problems of labeling failure and low detection limit of library construction methods, and achieve high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 3
[0081] Example 1 Establishment of three rounds of PCR amplification library construction method
[0082] This embodiment provides a high-sensitivity amplicon sequencing kit and usage method.
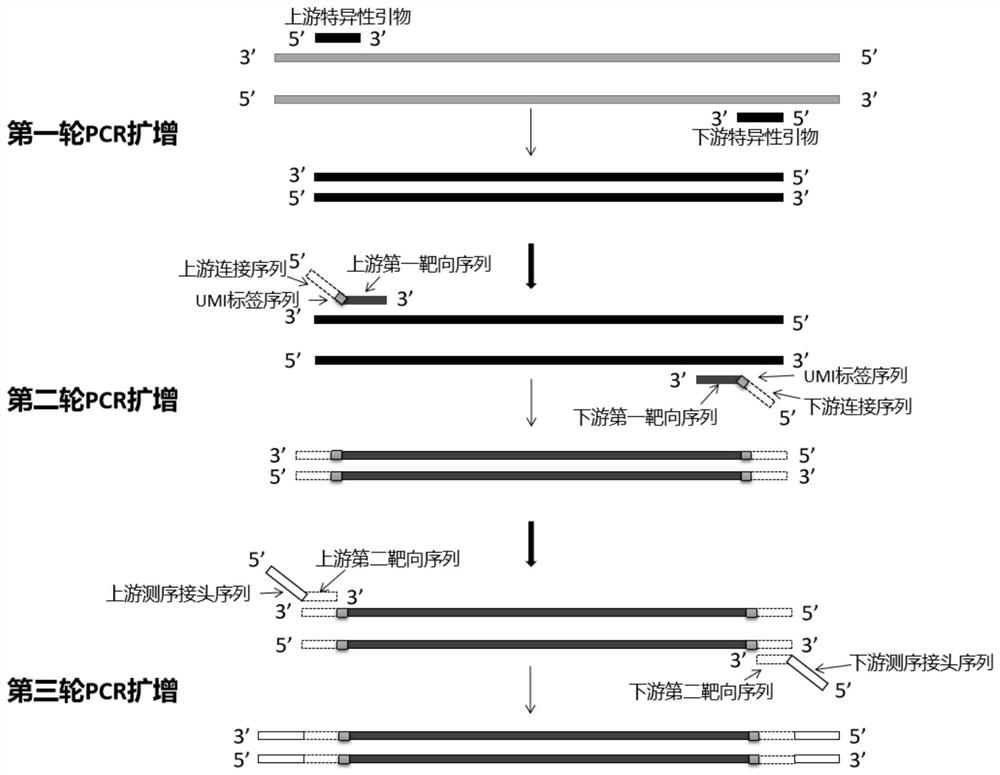
[0083] The kit includes three primer pairs: a specific primer pair for amplifying the target region; a primer pair for adding a specific molecular tag (Unique Molecular Identifier, UMI); a primer pair for adding sequencing adapters. Among them, the primers of the primer pair for adding UMI include the sequence of the amplification product of the connecting sequence, the UMI sequence and the target-specific primer pair from the 5' end to the 3' end; the primers of the primer pair for adding the sequencing adapter The sequence from the 5' end to the 3' end includes the sequence of the sequencing adapter sequence and the targeting linker sequence.
[0084] Utilize above-mentioned kit to build library, comprise the following steps (such as figure 1 shown):
[0085] (1) Obtain a DNA sampl...
Embodiment 2
[0089] Embodiment 2 Utilizes the method for building a database established in Embodiment 1 to build a database
[0090] The inventor obtained DNA samples from two individuals and performed whole exome sequencing (WES) respectively to obtain low frequency loci.
[0091] (1) The first round of PCR amplification
[0092] The variant sites shown in Table 1 were selected for targeted amplification and sequencing.
[0093] Table 1 Targeted amplification sequencing variant site information
[0094] Variation site number The chromosome where the mutation site is located start termination reference base Variant base 1 chr1 19018405 19018405 G A 2 chr1 43806166 43806166 G A 3 chr1 47746678 47746678 G C 4 chr1 65311262 65311262 G A 5 chr1 110883917 110883917 A G 6 chr1 157545384 157545384 T C
[0095] According to the mutation sites in Table 1, the target amplification region was selected and specific ...
Embodiment 3 Embodiment 2
[0160] Example 3 Sequencing and analysis of the library established in Example 2
[0161] The library constructed in Example 2 was sent to the Illumina NovaseqPE150 for sequencing and the number of 1M reads was analyzed.
[0162] Calculated by UMI2_PowerScan6 (www.genekras.com), the analysis results obtained:
[0163] As a comparison, the inventors used a two-step method to build a library (compared to Example 2, omitting the second step of using UMI tags to amplify, and the rest of the steps were the same), and using the same method to sequence the obtained library, analyze.
[0164] The library constructed in Example 2 was used for sequencing analysis, and the results are shown in Table 9.
[0165] Table 9 The sequencing analysis results of the library constructed in Example 1
[0166]
[0167] Note:
[0168] CHROM: Reference sequence name.
[0169] POS: the left-most position (1-base position) where the mutation is located, that is, the position of the first base of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com