Molecular marker closely linked with bacterial wilt resistance of cultivated peanuts and application of molecular marker
A peanut bacterial wilt and molecular marker technology, applied in the field of molecular biology, can solve the problem of inability to obtain molecular markers for bacterial wilt resistance molecular markers and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
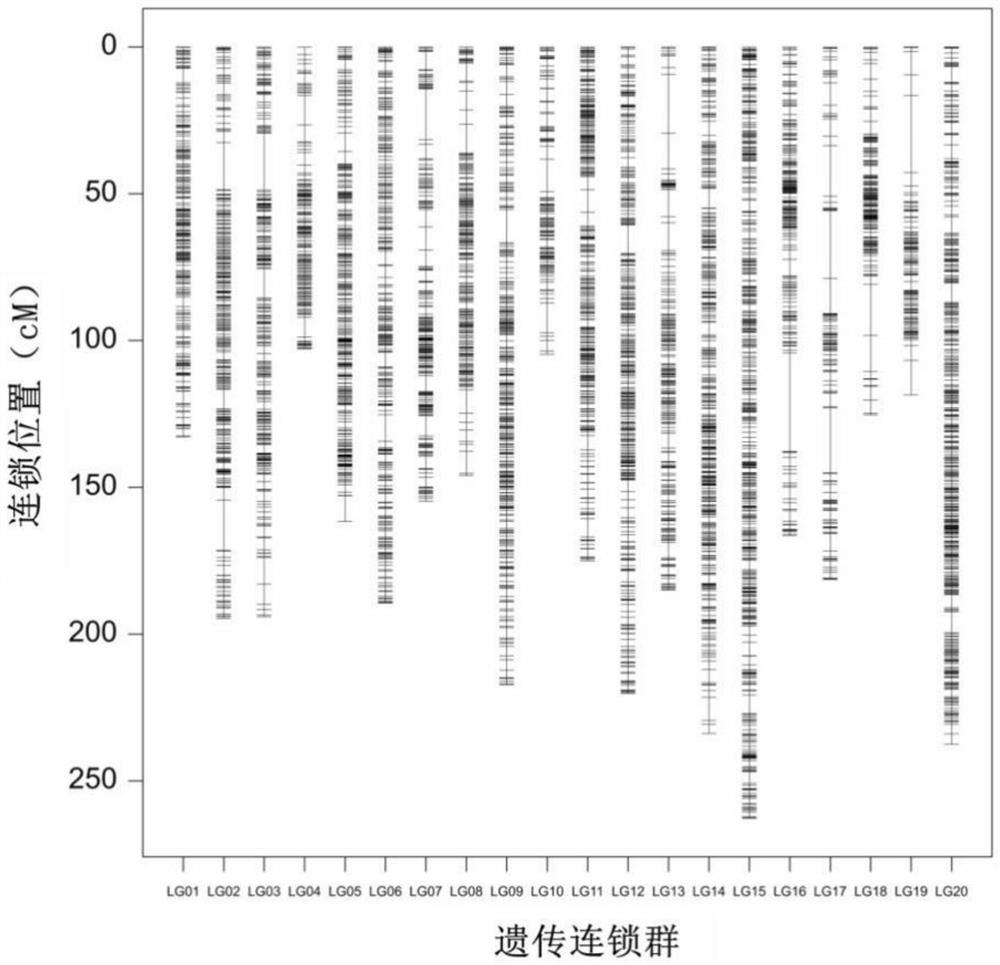
[0027] Example 1 Acquisition of main QTL for resistance to bacterial wilt of cultivar Peanut Yuanza 9102
[0028] (1) The recombinant inbred line populations of Yuanza 9102 and wt09-0023 (highly susceptible to bacterial wilt) were sequenced using simplified genome sequencing technology. The sequencing depth of the two parents was greater than 20×, and the sequencing depth of the progeny material was 5 ×. The genotyping data of the parents and offspring were obtained based on the genome information of cultivated peanut species (Arachis hypogaea.cv.Tifrunner V1.0). A total of more than 90,000 polymorphic SNPs with homozygous genotypes were obtained from the two parents.
[0029] According to the whole genome resequencing results of Yuanza 9102 and wt09-0023, KASP primers for 17 SNP sites were designed, and the genotyping data of the parents and offspring of the population were obtained through the genotyping platform. Filter out the SNP markers with a deletion rate of more tha...
Embodiment 2
[0032] Example 2 Identification of Molecular Markers Tightly Linked to the Major QTL for Bacterial Wilt Resistance in Yuanza 9102 Derivative Lines and Natural Populations
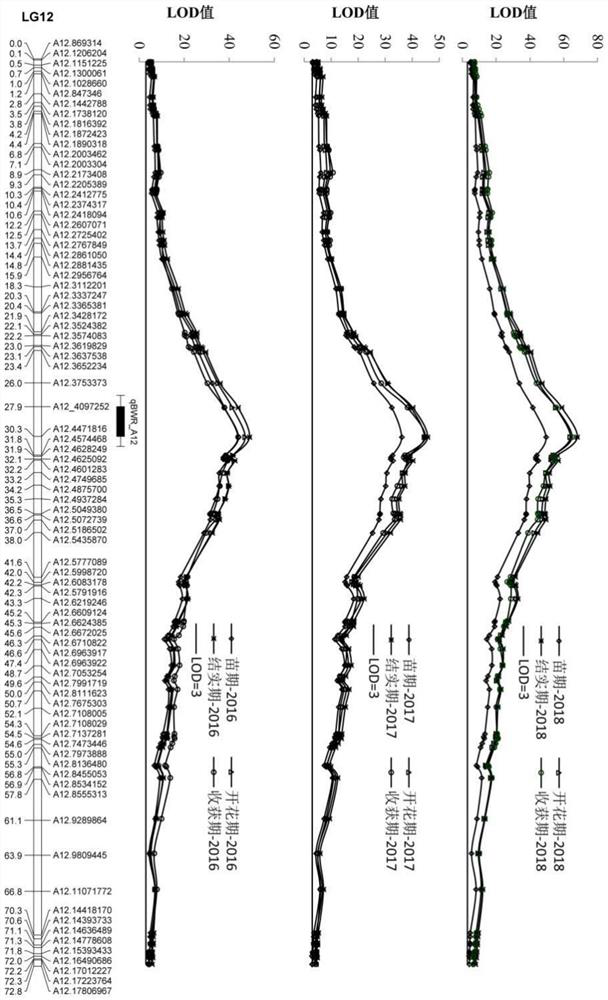
[0033](1) According to the genome sequence information of the SNP positions on the left and right flanks of the main QTL (qBWR_A12), 8 sets of KASP primers were developed (see Table 1), and these SNP sites were verified by the SNPLine genotyping platform (LGC) in Yuanza 9102 and wt09-0023 in recombinant inbred lines whose genotyping results were consistent with the sequencing results.
[0034] Table 1 KASP primer sequence information
[0035]
[0036] (2) Select 47 Yuanza 9102 derived line materials (see Table 2) and 317 natural population materials (see Table 3) to carry out resistance phenotype identification in the bacterial wilt disease nursery of Hezhou Peanut Comprehensive Experimental Station, Guangxi Province . The 47 materials derived from Yuanza 9102 are intermediate breeding materials from d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com