Method for diagnosing SARS-CoV-2 infection
A sars-cov-2, protein technology, applied in the field of SARS-CoV-2 infection diagnosis, can solve the problems of invasion, inconvenient sampling, poor diagnostic accuracy, etc., achieve high accuracy, save medical care costs, facilitate sampling and quick effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Collection and treatment of saliva samples
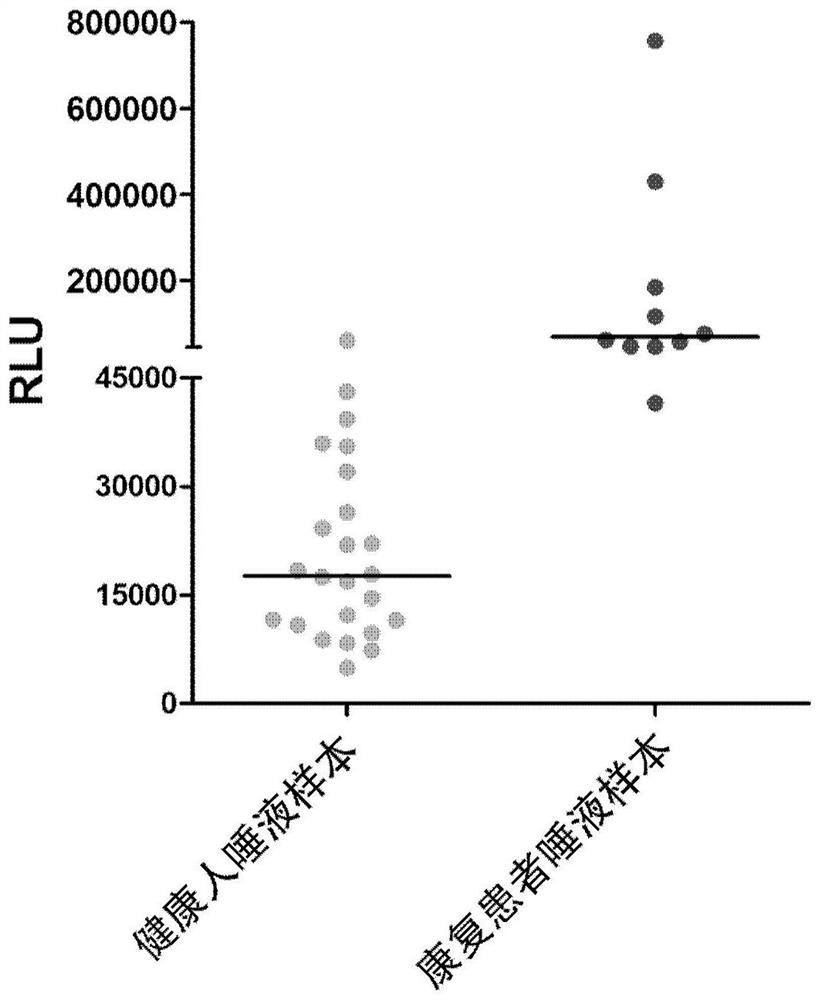
[0031] 1) Randomly select two healthy people, as well as four COVID-19 rehabilitation patients as the detected detectors 1, 2, 3, 4, 5, 6, and 6. Inform the detected person, it is not possible to perform diet, mouthwash, etc., can affect the saliva quality in at least 10 minutes before taking a sample. At the same time, the saliva collection container is issued to the detected, and the collection container opening is preferably greater than 5 cm and has a sealing cover.
[0032] 2) Clean the surface of the container on the surface of about 1 ml of saliva.
[0033] 3) Collect the sampled saliva samples of the detected, and use 75% alcohol to disinfect the outer surface of the vessel.
[0034] 4) -20 ° C Save, open in the biosafety cabinet during detection.
Embodiment 2
[0035] Example 2. SARS-COV-2 specific IgA in saliva samples using immunopolymerization method
[0036] 1) Expression of the recombinant binding region of purification of SARS-COV-2 ugly proteins (SEQUENCE ID: MT322424.1, specific nucleic acid sequence SEQ ID NO.1 See the last page of the specification), and coupled to CNBR-ACTIVATED Sepharose TM 4B Agarose beads (purchased from GE).
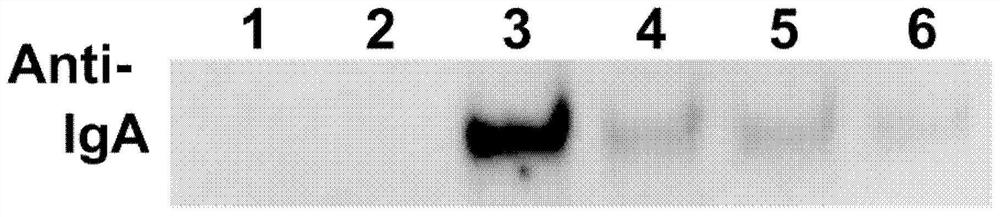
[0037] 2) Take the above 2 healthy people and 4 COVID-19 rehabilitation patients (i.e., detected 1, 2, 3, 4, 5, 6) each 1 ml of each of which were diluted with 4 ml of PBS and transferred to 10 ml of centrifuge tube, respectively. Each addition of 100 μL of the agarose beads of the coupling RBD prepared in the first embodiment of the present embodiment, mixing, and incubated at room temperature for 30 min.
[0038] 3) 1000g Centrifuge 1min and discard it. Then, 4 ml of PBS was added, mixed upside down 20 times to clean the beads.
[0039] 4) 1000g Centrifuge 1min, repeating this Example 2 Step 3) 4 t...
Embodiment 3
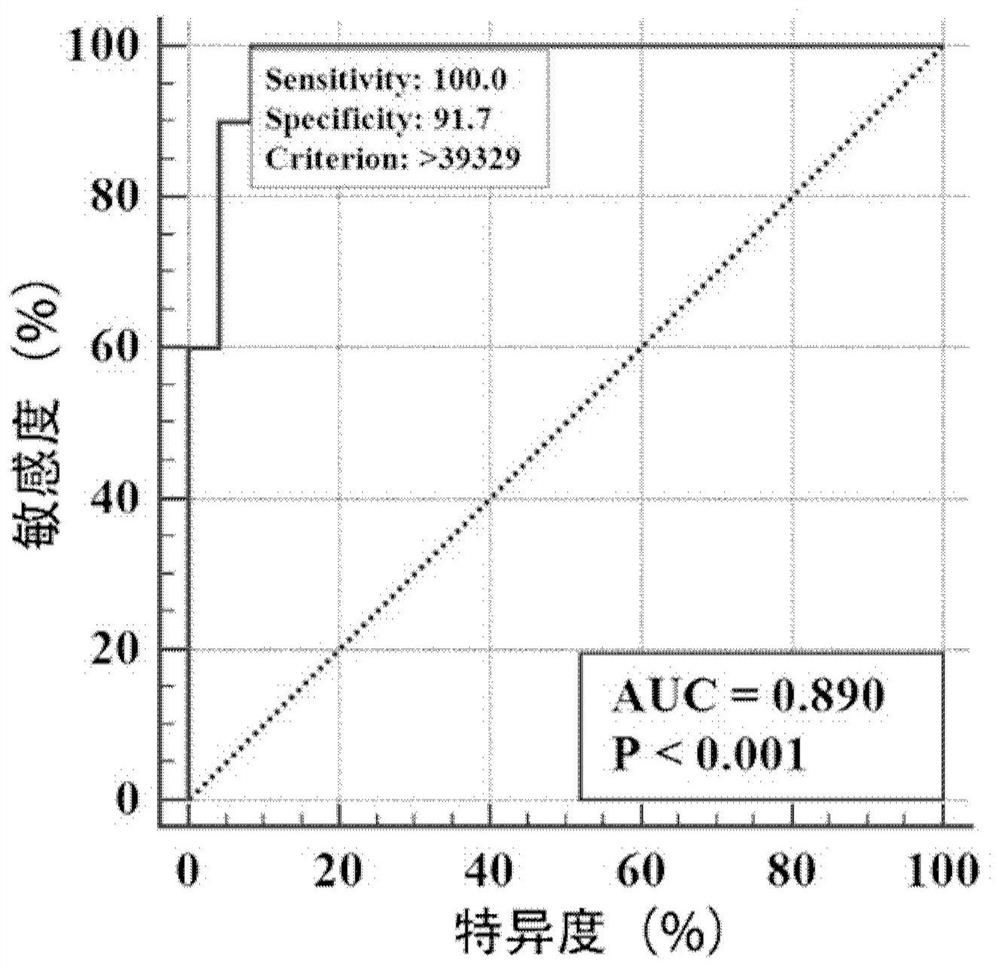
[0047] Example 3. The detection principle of SARS-COV-2 specific IgA utilization chemiluminescence method was detected by chemiluminescence method: the sample to be tested by saliva and the magnetic beads with SARS-COV-2 RBD After incubation, after the washing washing washed, the anti-human IgA antibody acridate syrids were added to incubate together, and after washing, the substrate fluid was added, and the luminescence reaction of the acridine ester was then detected. If there is a new coronavirus IgA antibody in the sample, a magnetic bead package can be formed to form a positive correlation between the luminescence intensity of the acridine ester with a new coronavirus IgA antibody. Relationship, the test results are expressed in the critical value index (COI).
[0048] The chemiluminescent detection of this embodiment is used in full automatic detection machine Kreser 1000, a yin-positive control, and is used to screen the correction.
[0049] The specific steps are:
[0050]...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com