Adenine base editor fusion protein without PAM limitation and application
A fusion protein and gene editing technology, applied in the field of biomedicine, can solve the problems of unresolved off-target problems, low editing efficiency, and restrictions on the application of base editors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0111] Example 1 Construction of base editor plasmid
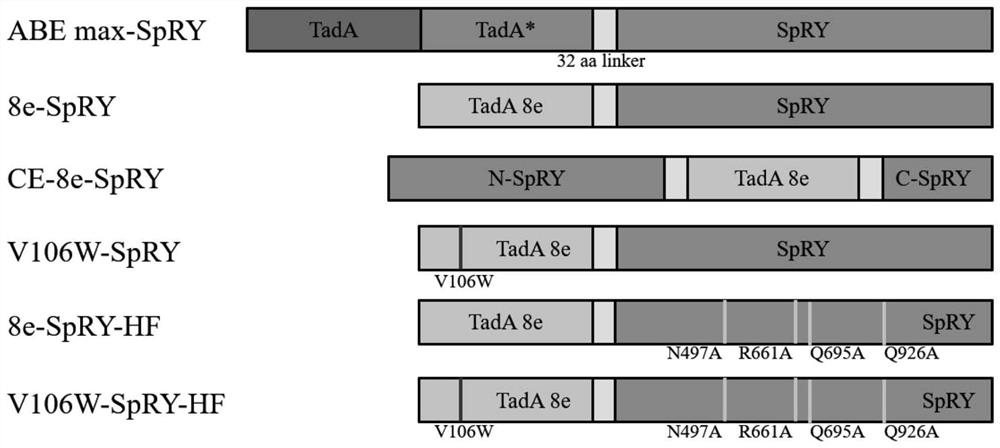
[0112]First construct 8e-SpRY and corresponding mutants. Refer to the instructions of ClonExpress MultiS One StepCloning Kit (Vazyme, C113-01) to design primers to amplify the TadA8e fragment in ABE8e (Addgene #138489), and replace the TadA dimer in ABEmax-SpRY (Addgene #140003) with TadA8e to construct 8e-SpRY plasmid.
[0113] First delete TadA8e in 8e-SpRY from the original position, and then replace the 1048th to 1063rd amino acids in SpRY D10A with TadA8e to construct the CE-8e-SpRY plasmid, and the sequence from the 5' end to the 3' end is SpRY (D10A) N-terminal, TadA8e and SpRY (D10A) C-terminal, wherein the nucleotide sequence of SpRY (D10A) N-terminal is as shown in SEQ ID NO: 2 (amino acid sequence is shown in SEQ ID NO: 1), the nucleotide of TadA8e The sequence is shown in SEQ ID NO: 4 (the amino acid sequence is shown in SEQ ID NO: 3), and the nucleotide sequence at the C-terminal of SpRY (D10A) is shown in S...
Embodiment 2
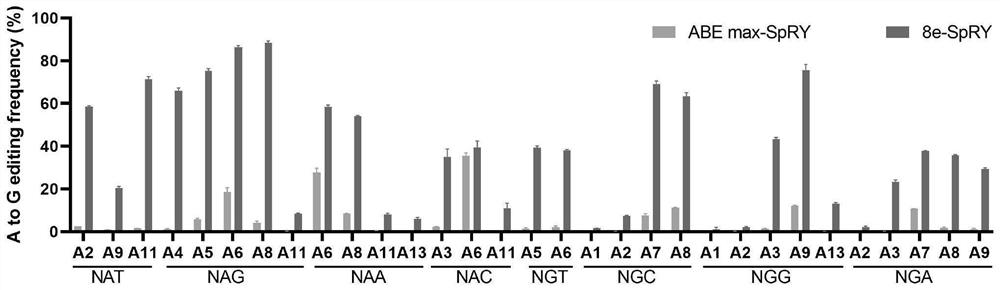
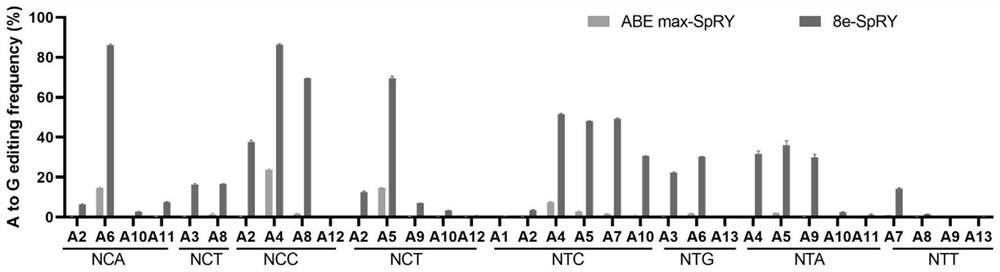
[0131] In this example, ABEmax-SpRY, 8e-SpRY and their mutants were used to edit endogenous sites in 293T cells.
[0132] 2.1 Construction of sgRNA plasmid
[0133] Referring to the human genome sequence, 48 sgRNAs were designed according to the PAM characteristics of SpRY nuclease, covering 16 different PAM sequences. The sgRNA sequence is shown in SEQ ID NO: 18-65. ACCG is added to the 5' end of the sgRNA sequence as the upstream sequence. The sgRNA AAAC is added to the 5' end of the reverse complementary sequence as the downstream sequence, and the upstream and downstream sequences are annealed after the oligo is synthesized (program: 95°C, 5min; 95°C-85°C at-2°C / s; 85°C-25°C at-0.1 ℃ / s; hold at 16℃) and then ligated with the pGL3-U6-sgRNA (Addgene #51133) vector digested with BsaI (NEB: R3733L). The enzyme digestion system is: pGL3-U6-sgRNA 2μg; CutSmart buffer (NEB: B7204S) 6μL; BsaI 1μL; ddH 2 Make up to 60 μL with O, digest overnight at 37°C. The ligation system is: ...
Embodiment 3
[0152] In this example, the RNA off-target situation of ABEmax-SpRY, 8e-SpRY and their mutants in 293T cells was compared.
[0153] 3.1 Construction of sgRNA
[0154] The sgRNA sequence used for RNA off-target detection is 5'-CTGGAACACAAAGCATAGAC-'3 (SEQ ID NO:66), which was constructed according to the plasmid construction method described in 2.1.
[0155] 3.2 Cell culture and transfection
[0156] Cell culture was carried out as described in 2.2. The day before transfection, 293T cells were used to spread a 6cm dish, so that the cell density at the time of transfection reached about 80%. The amount of plasmid transfected per dish is 4 μg of base editor plasmid and 2 μg of sgRNA plasmid. Dilute the plasmid in 250 μL of DMEM, and dilute 18 μL of EZ Trans cell transfection reagent (Shanghai Lee Kee Bio: AC04L092) in 250 μL of DMEM Finally, add the diluted EZ transfection reagent to the diluted plasmid, mix well and let stand at room temperature for 15 minutes. Add the DMEM m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com