Composition for detecting bladder cancer and kit and application thereof
A detection kit and technology for bladder cancer, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of high experimental cost, reduced specificity, multiple reagents, etc., and improve detection sensitivity , reduce the operation steps, reduce the effect of reagent consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1: Sample DNA Extraction and Bisulfite Conversion
[0068]1. Sample processing and DNA extraction
[0069] 1) Sample pretreatment:
[0070] Tissue and cell samples do not need to be pretreated, and the pretreatment of urine samples is as follows: take 80mL of morning urine in a 100mL centrifuge tube, centrifuge at 4200rpm for 10min, discard the supernatant, add 10mL of PBS to mix and wash the cell sediment, centrifuge at 4200rpm for 5min, discard part of the supernatant Only keep about 1mL supernatant, mix well and transfer to a 1.5ml sterile centrifuge tube.
[0071] 2) DNA extraction:
[0072] Extract bladder exfoliated cell DNA by using nucleic acid extraction or purification reagent (general type) produced by Anhui Dajian Medical Technology Co., Ltd. The specific steps are as follows:
[0073] (a) Add 200 μL Lysis Solution I and 30 μL Proteinase K to the sample to be tested, mix thoroughly, and incubate at 60°C for 2 hours until the sample is completely l...
Embodiment 2
[0091] Example 2: Screening of bladder cancer hypermethylation candidate genes and specific primers and probes
[0092] 1. Screening of candidate genes for urinary hypermethylation in patients with bladder cancer
[0093] Comprehensive analysis of literature research results, TCGA methylation chip database and transcriptome sequencing expression profiles, screening of methylation sites with significant differences, and through multiple data filtering analysis, the final screening determined that ECRG4, TMEFF2, and TWIST1 are candidates for bladder cancer hypermethylation Gene.
[0094] 2. Screening of primer-probe combinations for methylation detection of bladder cancer
[0095] 1) Screening of specific primers and probes:
[0096] According to the nucleic acid sequences of ECRG4, TMEFF2, and TWIST1 mentioned above, methylation primers and probes were designed on the Methyl primer Expressv1.0 software. After repeated design and deliberation by the applicant, PCR probes and p...
Embodiment 3
[0102] Example 3: Methylation fluorescence quantitative PCR amplification detection of bladder cancer hypermethylation candidate genes
[0103] 1. The reaction system of methylation fluorescent quantitative PCR is as follows: 10 μL of 2×methylation PCR reaction master mix, 0.1 μL of 10 μM GAPDH primers and probes, 0.5 μL of each primer in the 10 μM primer-probe combination, and 0.5 μL of each probe. 0.2 μL, 5 μL Bis-DNA, and rehydrate to 20 μL.
[0104] 2. Reaction conditions for methylation fluorescent quantitative PCR
[0105]
[0106]
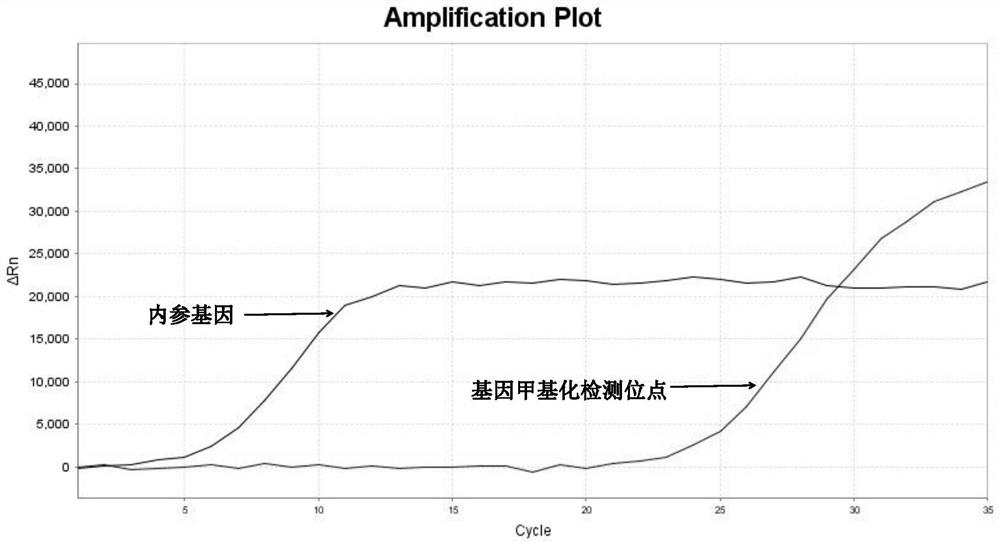
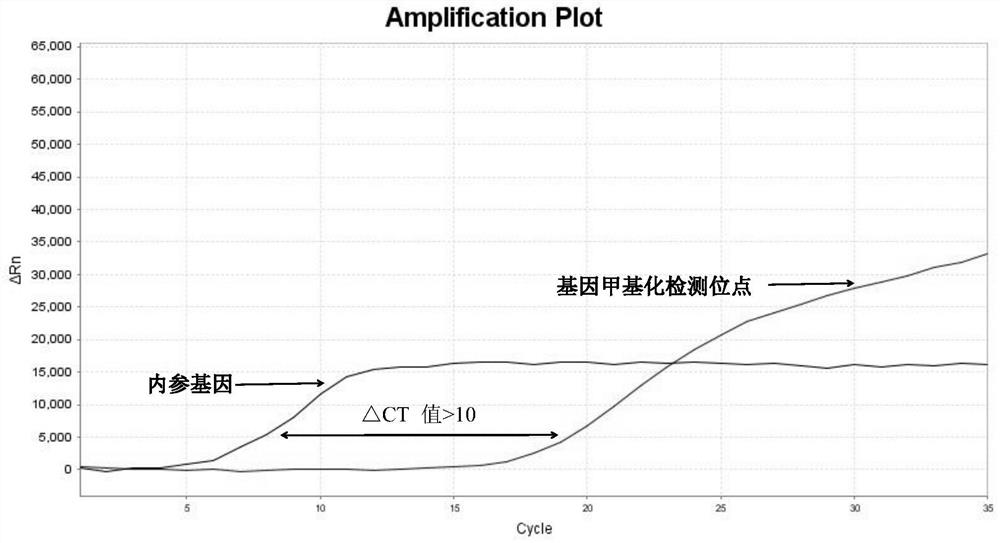
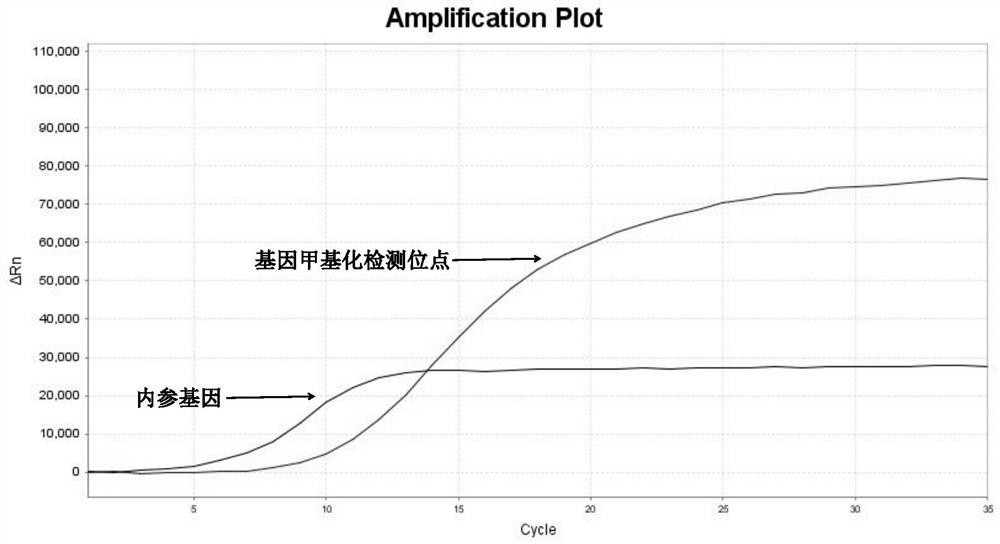
[0107] 3. Interpretation of detection results of methylation fluorescent quantitative PCR
[0108] 1) Threshold value setting: It can be output automatically according to the instrument, or manually adjust the baseline according to the instrument’s instructions, set the threshold value at the linear part of the logarithmic graph of the fluorescence value, export the data from the software and read the CT value.
[0109] 2) Judgment o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com