Cervical cancer gene methylation detection primer probe combination and kit and application thereof
A technology of detection kits and primer probes, applied in the biological field, can solve the problems of high experimental cost, limited diagnostic effect, and reduced specificity, and achieve the effect of reducing labor costs, reducing consumable costs, and reducing operating steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1: Sample DNA Extraction and Bisulfite Conversion
[0067] 1. Processing of cervical exfoliated cell samples and DNA extraction
[0068] 1) Sample collection:
[0069] The operation of collecting cervical exfoliated cell samples is as follows: the medical staff first exposes the cervix with a speculum, and wipes off the excessive secretion of the cervix with a cotton swab. Place the cervical brush on the cervix and rotate it 5 times in one direction to obtain a sufficient sample of epithelial cells, then put the head of the cervical brush into the sample tube containing the cell preservation solution, and place the cervical brush handle along the crease of the brush handle. Break off, leave the brush head in the sample tube, tighten the tube cap, and mark the sample
[0070] 2) DNA extraction:
[0071] Extract the DNA of cervical exfoliated cells by using the nucleic acid extraction or purification reagent (general type) produced by Anhui Dajian Medical Techn...
Embodiment 2
[0091] Example 2: Screening of cervical cancer hypermethylation candidate genes and specific primers and probes
[0092] 1. Screening of candidate genes for hypermethylation in cervical exfoliated cells of patients with cervical cancer
[0093] Comprehensive analysis of literature research results, TCGA methylation chip database and transcriptome sequencing expression profiles, screening of methylation sites with significant differences, and through multiple data filtering analysis, final screening and determination of SOX1, AJAP1, and ZNF671 as candidates for cervical cancer hypermethylation Gene.
[0094] 2. Screening of primer-probe combinations for cervical cancer methylation detection
[0095] 1) Screening of specific primers and probes:
[0096] According to the nucleic acid sequences of SOX1, AJAP1, and ZNF671 mentioned above, methylation primers and probes were designed on the Methyl primer Express v1.0 software. After repeated design and deliberation by the applican...
Embodiment 3
[0101] Example 3: Methylation fluorescence quantitative PCR amplification detection of cervical cancer hypermethylation candidate genes
[0102] 1. The reaction system of methylation fluorescent quantitative PCR is as follows: 7.5 μL of 2×PCR reaction master mix, 0.1 μL of 10 μM GAPDH primers and probes, 0.5 μL of each primer and 0.2 μL of probes in the 10 μM above primer-probe combination , 3 μL Bis-DNA, and rehydrate to 15 μL.
[0103] 2. Reaction conditions for methylation fluorescent quantitative PCR
[0104]
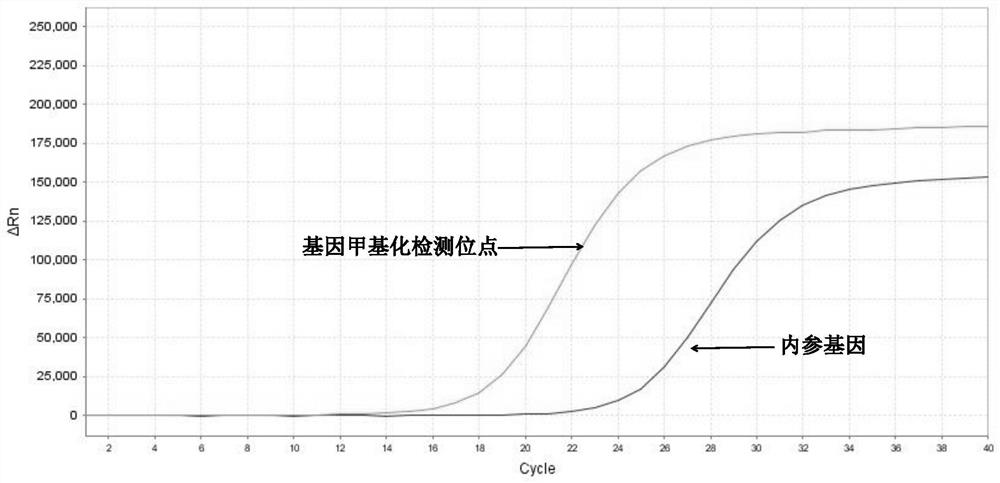
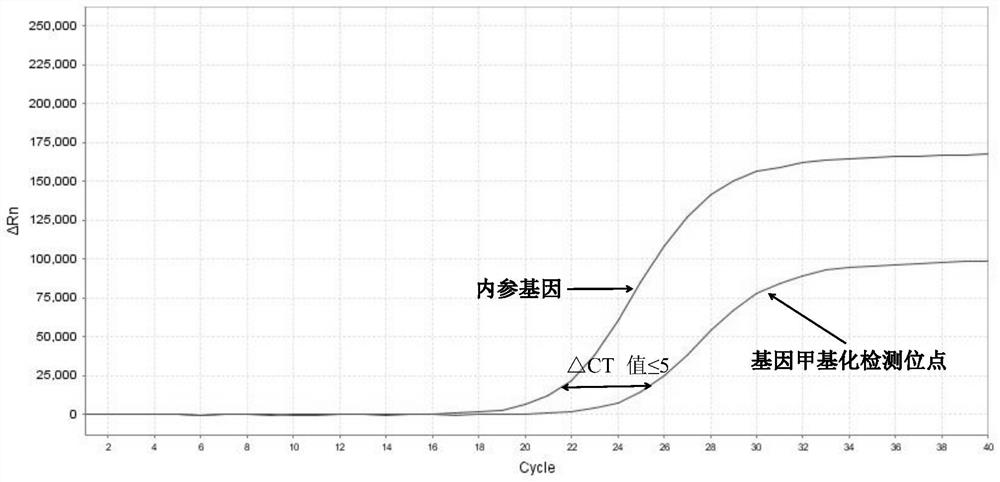
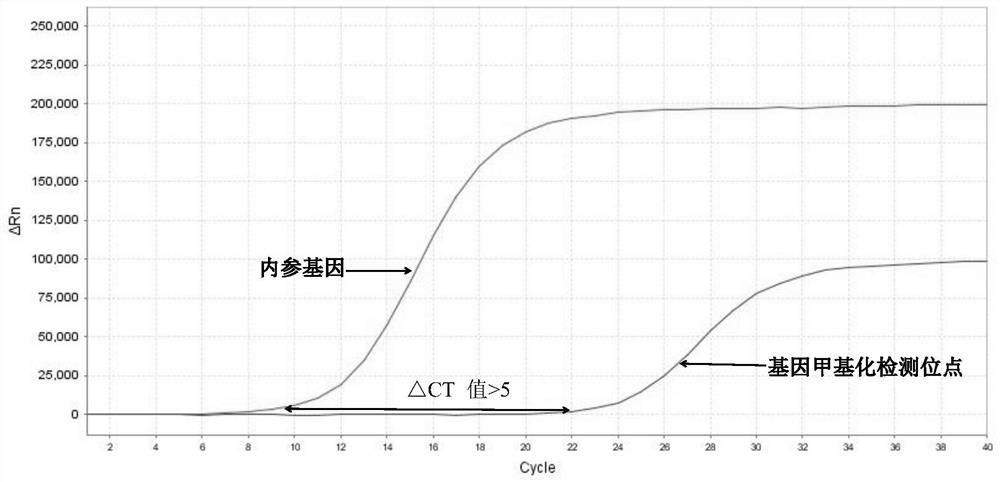
[0105] 3. Interpretation of detection results of methylation fluorescent quantitative PCR
[0106] 1) Threshold value setting: It can be output automatically according to the instrument, or manually adjust the baseline according to the instrument’s instructions, set the threshold value at the linear part of the logarithmic graph of the fluorescence value, export the data from the software and read the CT value.
[0107] 2) Judgment of the effectiveness of the k...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com