RNA-seq differential expression gene determination method and application
A technology of differentially expressed genes and determination methods, applied in the field of single-cell organisms, can solve the problems of selecting differentially expressed genes, lack of uniform standards for differentially expressed genes, etc., and achieve the effect of reducing errors and human intervention errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] The following describes several preferred embodiments of the present invention with reference to the accompanying drawings, so as to make the technical content clearer and easier to understand. The present invention can be embodied in many different forms of embodiments, and the protection scope of the present invention is not limited to the embodiments mentioned herein.
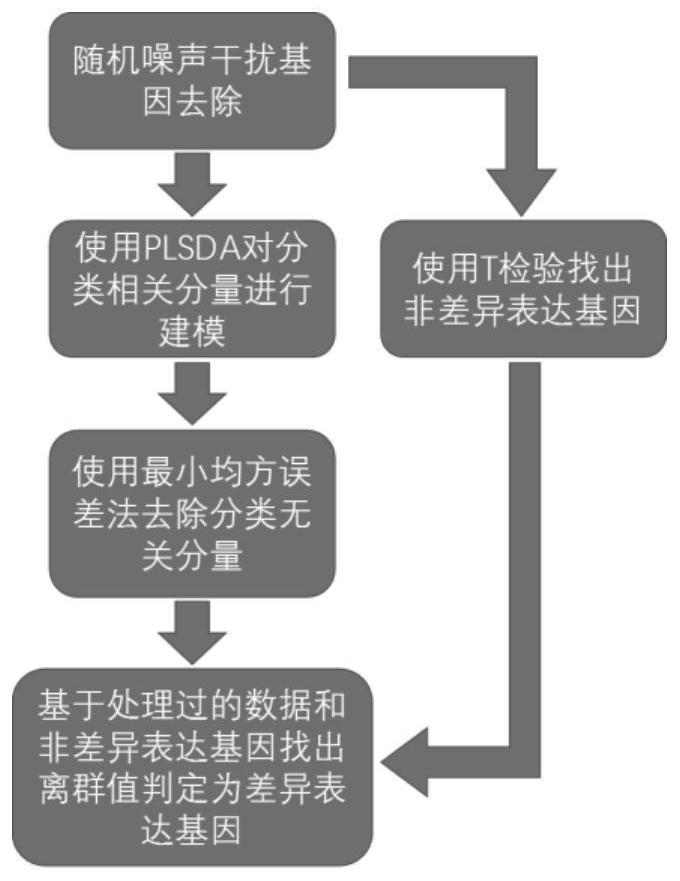
[0032] Such as figure 1 As shown in the algorithm operation flow chart, the process of the dynamic modeling algorithm for single-cell transcriptome data is described in detail, as follows:
[0033] Step 1. Random noise interference gene removal: Merge cells from two classes, X and Y, together. Based on the average expression level μ and variance σ of each gene 2 Relationship: σ 2 / μ 2 = a0+al / μ, calculate the predicted value σ of the variance of each gene 1 2 . If the true variance value σ of the gene 2 1 2 , the gene is considered to be a noise pollution gene.
[0034] Step 2. Use t-test to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com