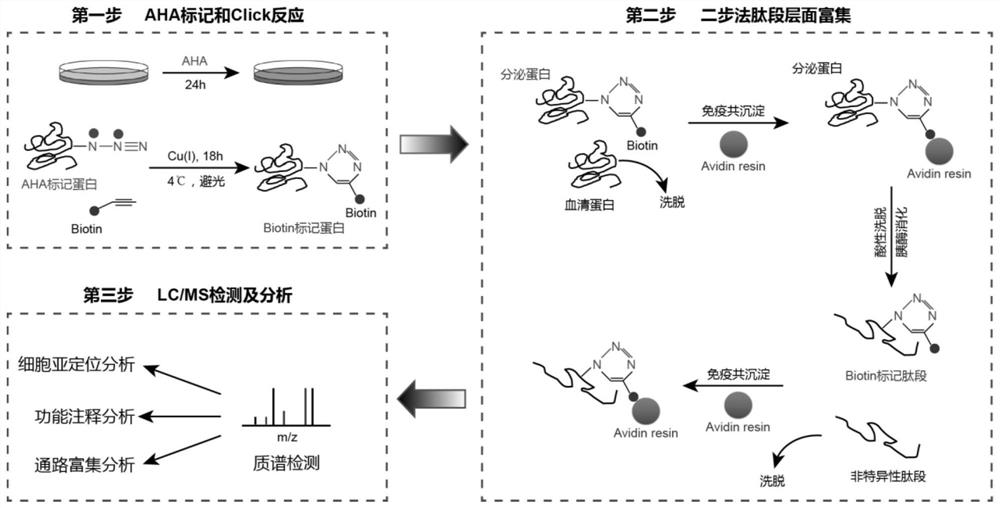
High-specificity cell secreted protein enrichment method
A technology for cell secretion and secretion of proteins, which is applied in the field of proteomics and can solve problems such as the influence of secretome, cell death, and cell state.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
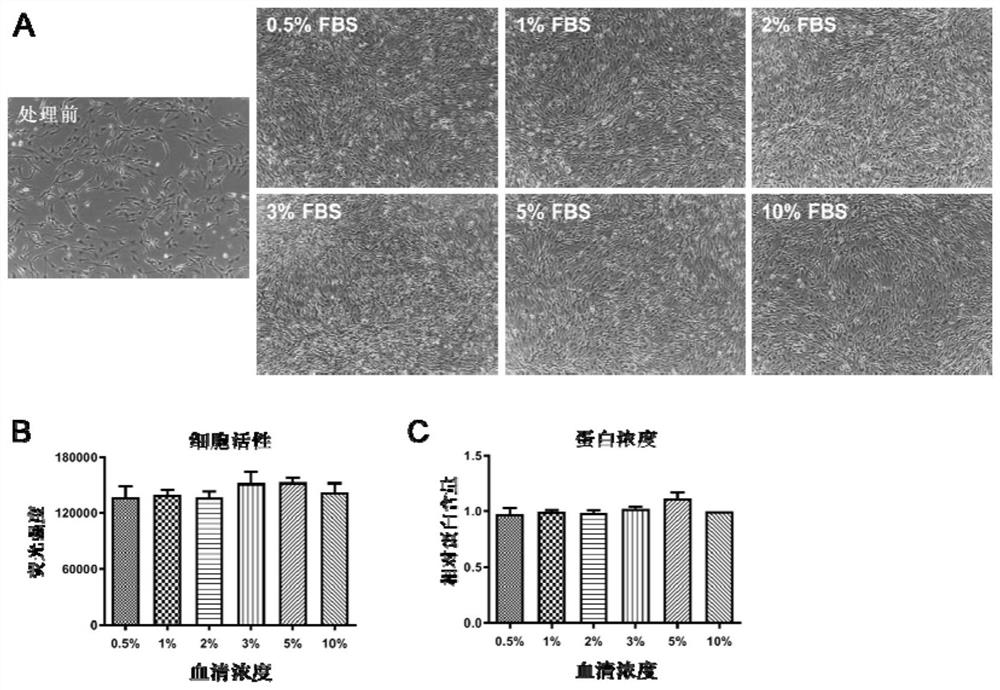
[0140] Example 1: Exploration of low serum culture concentration
[0141] Since the high concentration of serum protein in the culture medium will affect the efficiency of the click chemical reaction, the present invention first explored the lowest serum concentration that cells can tolerate during cell culture. In this embodiment, five serum concentration groups of 0.5%, 1%, 2%, 3 and 5% were set up for exploration.
[0142] First let the cells grow to a density of 70-80% in ordinary medium (containing 10% FBS), and then change the medium containing the above concentrations of serum to culture the cells for 24 hours. Cells were photographed 24 hours later using a brightfield microscope.
[0143] The results showed that during the 24-hour culture period, there was no significant difference in the growth rate of cells under each serum concentration, and the cell density after 24 hours was basically the same ( figure 2 A).
[0144] Next, the cell viability of each group was ...
Embodiment 2
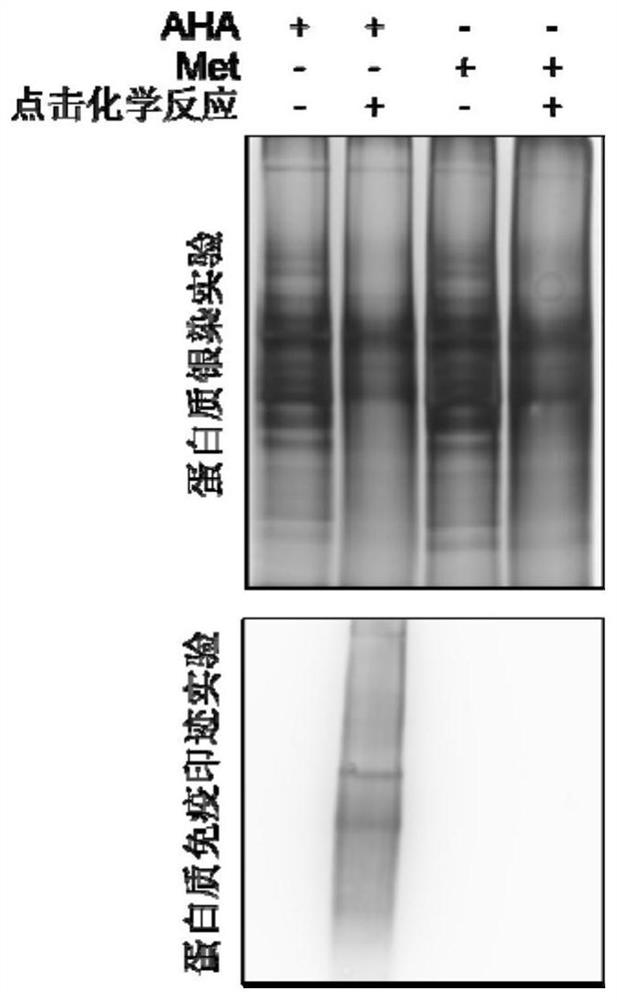
[0148] Example 2: AHA labeling and sample collection
[0149] The present invention uses a healthy human embryonic stem cell (hESC)-H1 (purchased from Wicell) induced astrocytes to enrich secreted proteins. The source and background of the cell line are clear , and used after astrocyte-specific protein expression detection and mycoplasma detection.
[0150] Usually, when the cells grow to 80-90% density in ordinary medium, they can be used for AHA labeling. The ordinary medium formula here is determined according to the specific cell type. The medium formula used in this method is shown in the table 1 and 2 are shown. There is a positive correlation between the initial cell volume when collecting secreted proteins and the final protein volume that can be detected. The specific initial cell volume needs to be determined according to the cell type and the secretory capacity of the cells. It needs to be determined in advance according to the state of the cells used. Confirmed b...
Embodiment 3
[0161] Example 3: Click Chemical Reaction
[0162] The sample after the above treatment can be sequentially added with triazole ligand, biotin-tetrapolyethylene glycol-alkyne (purchased from Sigma Company), and CuBr (purchased from Sigma Company) for click chemical reaction. The specific reaction system is shown in the table 4.
[0163] Table 4: Click chemistry reaction system (taking 1ml system as an example)
[0164]
[0165] It should be pointed out here that the triazole ligand used in this method was synthesized by referring to the literature and dissolved in DMSO (purchased from Sigma). The CAS number of the triazole ligand is 510758-28-8 and can also be purchased through the company. Meanwhile, the method is catalyzed by monovalent copper (CuBr), but the method is not limited to monovalent copper, divalent copper can also catalyze the reaction. And it should be noted that CuBr is easy to decompose, and it needs to be prepared and used before each reaction. In add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com