Probe, primer and kit for detecting ACE2 gene polymorphism
A gene polymorphism and kit technology, applied in the field of molecular biology, can solve the problems of high cost, time-consuming and laborious, unsuitable detection, etc., and achieve the effect of simple operation, improved curative effect, and rapid screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Probe and Primer Design
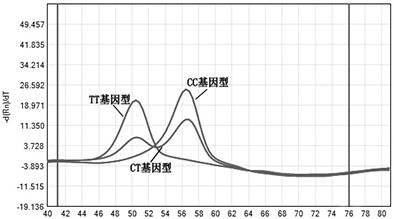
[0026] The present invention designs probe and primer sequences for the ACE2 SNP site. The specific principle is to use the conformational change of the fluorescent probe and the target sequence to release the fluorescent dye, and to judge the genotyping result according to the peak maps and Tm values at different temperatures after hybridization. In the absence of target DNA, the fluorophore and quencher can be stably combined, and no fluorescent signal can be detected; when there is target DNA, the structure of the fluorescently labeled probe is destroyed, and the fluorophore and quencher When the quenching groups are separated from each other, the fluorescent signal can be detected.
[0027] Design probes and primers so that when the template does not exist at the annealing temperature, the probe is in a stem-loop state, including the loop sequence and its reverse complementary stem sequences on both sides, with a total length o...
Embodiment 2
[0034] Example 2 Detection of different genotype standards
[0035] 1. Use the plasmid ACE2 to construct and prepare the wild-type standard plasmid and mutant standard plasmid containing the rs2285666 site of the target gene (the source of the plasmid and the synthesis of the plasmid containing the target gene are synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.) . The accuracy of the sequence was confirmed by sanger sequencing. The genotype of the wild-type standard plasmid rs2285666 was CC; the genotype of the mutant standard plasmid rs2285666 was TT. The standard plasmid DNA concentration was normalized to 10 ng / ul.
[0036] 2. Use the probes and primers in Example 1.
[0037] 3. PCR reaction system:
[0038] 1) Add 7.5 ul of PCR Mix, 0.5 uM of primer 1 solution, 0.5 uM of primer 2 solution, and 0.1 uM of probe to each PCR reaction well in turn, and then add wild-type standard plasmid to 3 different PCR reaction wells respectively DNA, mutant standard plasmid D...
Embodiment 3
[0041] Example 3 Detection method performance analysis experiment
[0042] 1. Precision test
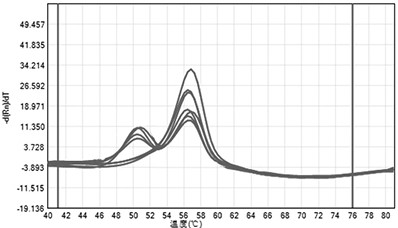
[0043]The wild-type standard plasmid DNA and mixed-type DNA (rs2285666 plasmid source, and the synthesis of the plasmid containing the target gene were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd. The wild-type standard plasmid and mutant standard plasmid were according to 1: 1 ratio of mixing), each of which was tested 3 times a day for 5 consecutive days. The amplification reaction procedure adopted the method in Example 2, and the results were shown in figure 2 . It shows that the fluorescence PCR amplification reaction of the present invention has good repeatability (the coincidence rate is greater than 95%, and the coefficient of variation CV of the Ct value of the detection result is less than 5%).
[0044] 2. Coincidence rate experiment
[0045] Twenty DNA samples from healthy volunteers in Shanghai were selected, and the method of Example 2 was used to detect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com