Linked molecular marker combination for major sites of sucrose content of peanuts, primer composition, identification method and application of linked molecular marker combination and primer composition
A primer composition and molecular marker technology, applied in biochemical equipment and methods, microbial measurement/testing, recombinant DNA technology, etc., can solve problems such as limiting the progress of high-sucrose sweet peanuts
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1. Acquisition of Peanut Sucrose Content-Linked Molecular Markers
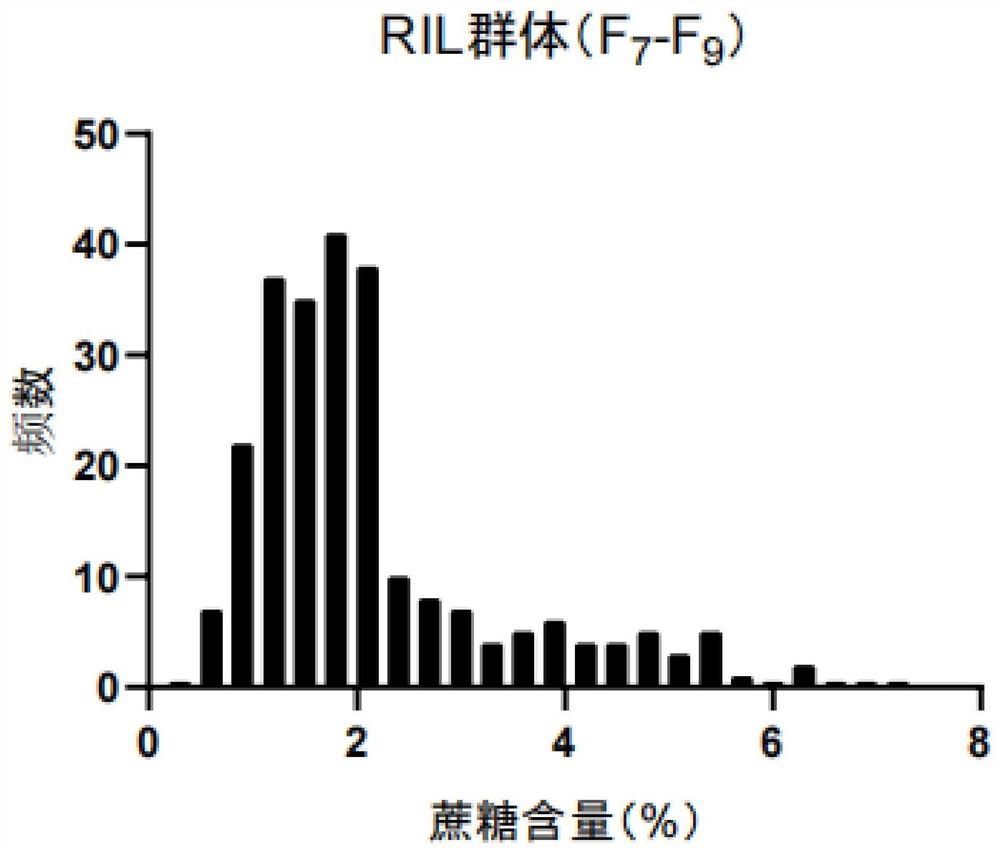
[0082] 1. Test materials: The peanut variety "Zhonghua 16" was used as the female parent, and the germplasm resource material "Rwanda Peanut" was used as the male parent for hybridization, and a recombinant inbred line containing 242 lines was constructed by the single-seed method ( RIL) Group F 7 (2017), F 8 (2018), F 9 (2019) Generation.
[0083]2. Phenotype identification: From 2017 to 2019, "Zhonghua 16", "Rwanda Peanut" and F 7 (2017), F 8 (2018) and F 9 (2019) Generation of 242 strains from the RIL population. A completely randomized block design was used with at least two repetitions. Every time is to repeat each strain and plant 1 row of 12 plants, the row spacing is 30 centimeters, and the plant spacing is 20 centimeters. Adopt standard field management methods to harvest mature pods of 8-10 typical plants with normal growth. Select 15 to 20 plump, mature and non-spotted kernel...
Embodiment 2
[0093] Example 2. Verification of Peanut Sucrose Content-Linked Molecular Markers
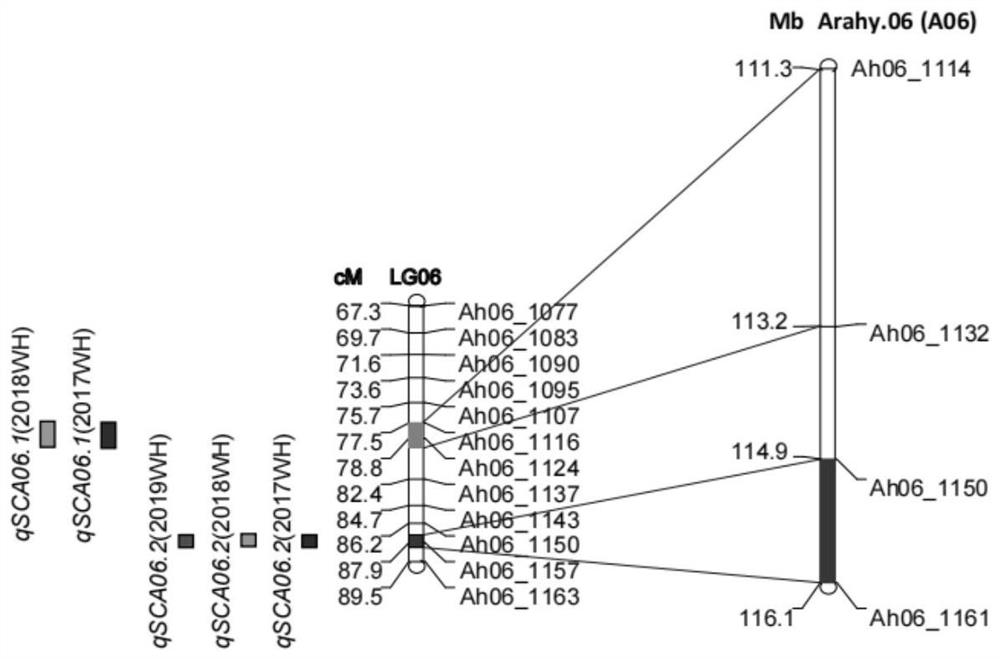
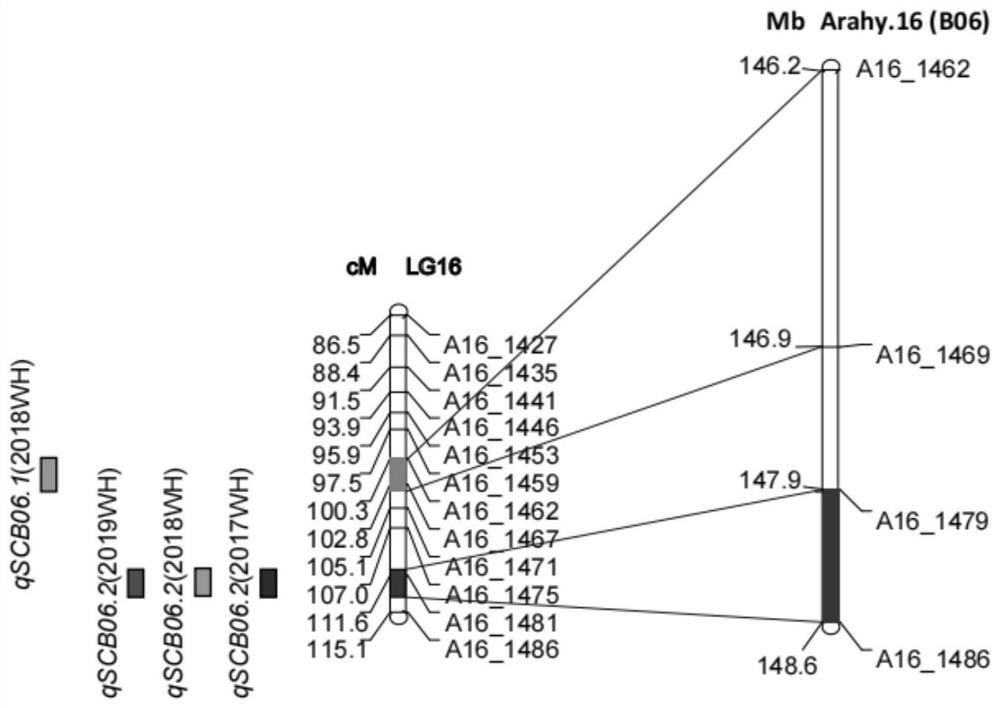
[0094] Using the three molecular markers closely linked to the main QTL site qSCA06.2 for peanut sucrose content and two molecular markers closely linked to the main QTL site qSCB06.2 for peanut sucrose content obtained in Example 1, the pair "Zhonghua 16" and 242 F's of "Rwandan Peanuts" 10 (2021) strains were PCR amplified. Using the InDel marker SCM06-11, SCM06-12 or SCM06-13 primer pair to amplify the same band pattern as the female parent "Zhonghua 16", denoted as aa, and amplify the same band pattern as the male parent "Rwanda Peanut" All are recorded as AA (Advanced Allele). Analyzing the phenotypic data of the RIL population (2017-2019), it was found that the average sucrose content of kernels of aa genotype materials was 1.58±0.03%, and the average kernel sucrose content of AA genotype materials was 2.69±0.04%. The three-year data showed that having the qSCA06.2 excellent allele cou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com