H4 nucleic acid aptamer and detection kit for detecting GBM cells and application of H4 nucleic acid aptamer and detection kit
A nucleic acid aptamer and detection kit technology, applied in the field of analysis and detection, can solve the problems of insufficient targeting and inability to accurately navigate the border of glioma, achieve good targeting, avoid interference from false positive signals, Good biocompatibility effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
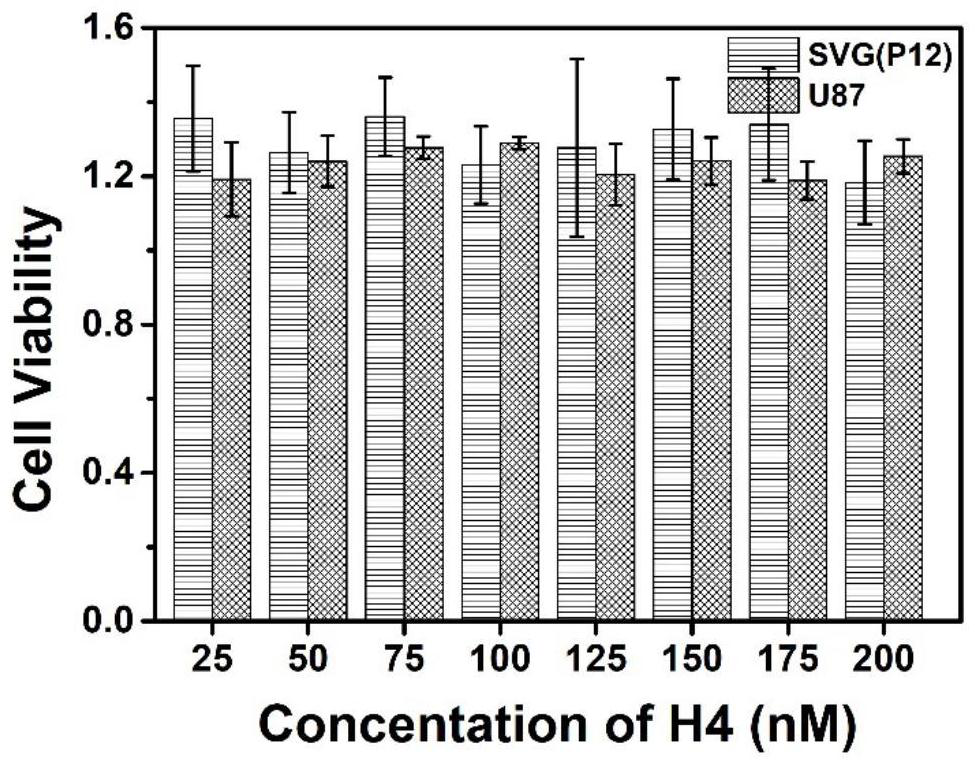
[0044] Detection of SVG(P12) glial cells
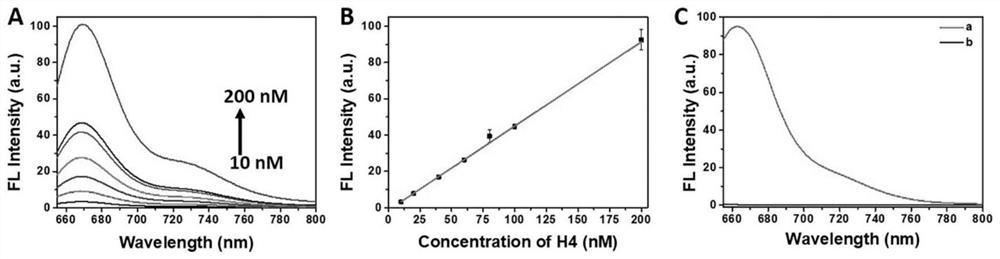
[0045] Select the SEQ ID NO.10 sequence of the 5' end and the 3' end to connect the fluorescent group Cy5 and the quenching group BHQ2 respectively (synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.), and the obtained powdery DNA H4-10 (SEQ ID NO.10) was centrifuged at 4000rpm for 60s, and 10μL of deionized water was added to make a stock solution DNA with a final concentration of 100μM, and then 90μL of buffer solution (5mM Tris-HCl, 250mM NaCl, 50mM MgCl 2 ) was further diluted to a final concentration of 10 μM stock solution DNA, fully shaken and mixed, heated to 95 ° C in a metal bath, kept for 5 min, and then slowly cooled to room temperature, and DNA with a hairpin structure was obtained through the above annealing steps. Such as figure 1 In (A) when the hairpin structure is not formed, the concentration of the aptamer ([aptamer]) and 666nm (F 666nm The fluorescence intensity at ) is positively correlated, and the func...
Embodiment 2
[0051] Detection of U87 glioma cells
[0052] Using the synthetic method of Example 1, the obtained powdered H4-10 (SEQ ID NO.10) DNA was centrifuged at 4000rpm for 60s, and 10 μL of deionized water was added to prepare a stock solution DNA with a final concentration of 100 μM, and then buffer solution (5 mM Tris -HCl, 250mM NaCl, 50mM MgCl 2 ) was further diluted to a final concentration of 10 μM stock solution DNA, fully shaken and mixed, heated to 95 ° C in a metal bath, kept for 5 min, and then slowly cooled to room temperature, and DNA with a hairpin structure was obtained through the above annealing steps.
[0053] Take 10 μL of the DNA solution of the above-mentioned hairpin structure and dissolve it in 900 μL DMEM, and co-incubate with U87 cells in a confocal glass-bottom dish at 37°C for 20 min.
[0054] Discard the supernatant, wash twice with PBS, fix with 4% paraformaldehyde for 20 min, wash away the 4% paraformaldehyde with PBS, and add 50 μL of cell nucleus dye ...
Embodiment 3
[0057] Detection of LN229 glioma cells
[0058] Using the synthetic method of Example 1, the obtained powdered H4-10 (SEQ ID NO.10) DNA was centrifuged at 4000rpm for 60s, and 10 μL of deionized water was added to prepare a stock solution DNA with a final concentration of 100 μM, and then a buffer solution (5 mM Tris-HCl, 250mM NaCl, 50mM MgCl 2 ) was further diluted to a final concentration of 10 μM stock solution DNA, fully shaken and mixed, heated to 95 ° C in a metal bath, kept for 5 min, and then slowly cooled to room temperature, and DNA with a hairpin structure was obtained through the above annealing steps.
[0059] Take 10 μL of the DNA solution of the above-mentioned hairpin structure and dissolve it in 900 μL DMEM, and co-incubate with LN229 cells in a confocal glass-bottom dish at 37°C for 20 min.
[0060] Discard the supernatant, wash twice with PBS, fix with 4% paraformaldehyde for 20 min, wash away the 4% paraformaldehyde with PBS, and add 50 μL of cell nucleus...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com