Genomic profiling: repid method for testing complex biological sample for presence of many types of organisms
A biological sample and genome technology, applied in the field of obtaining genetic information, can solve the problems of quantitative amplification methods that are difficult to design correctly and have no repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 5
[0151] Each of these steps is described in more detail below.
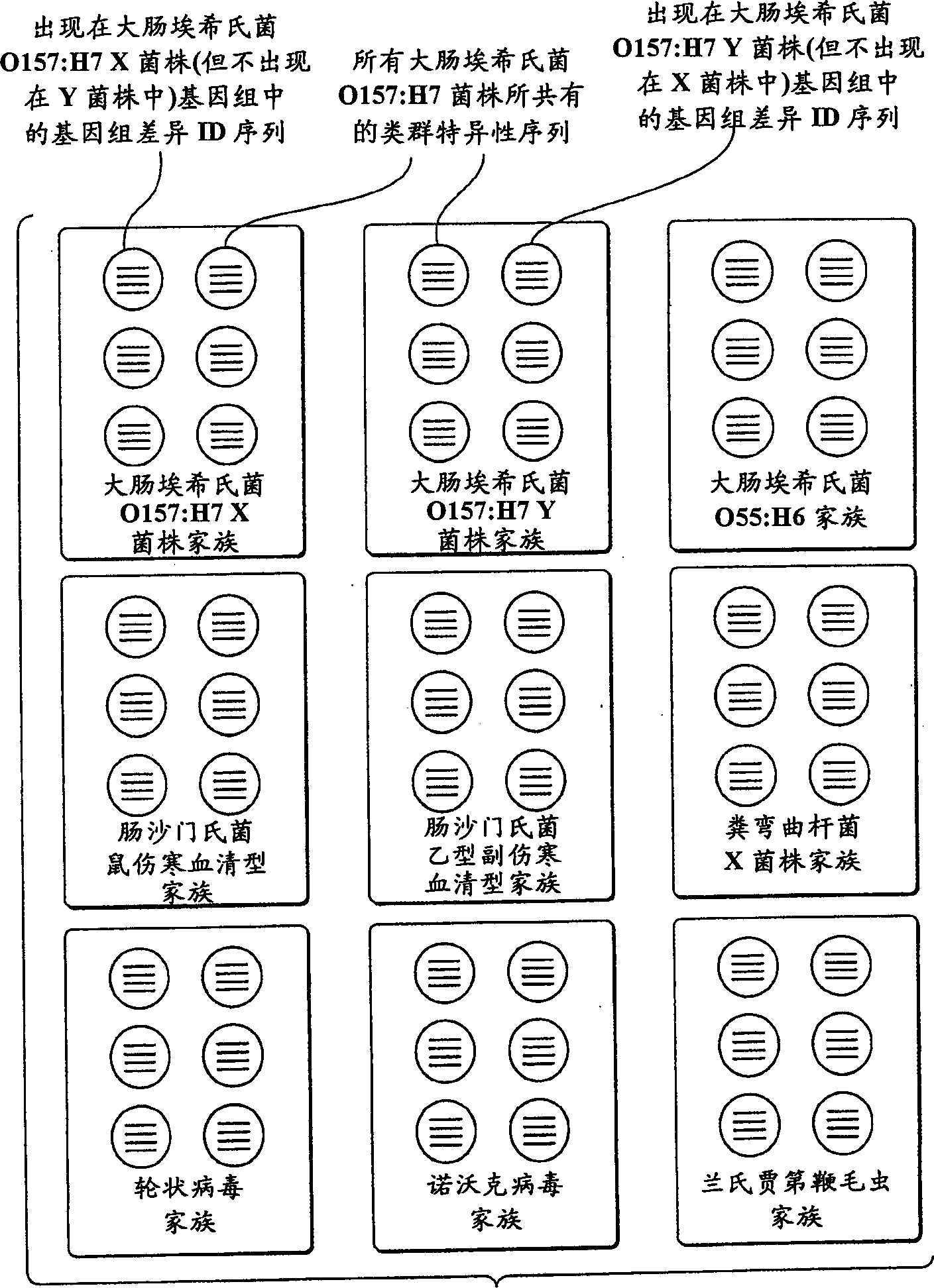
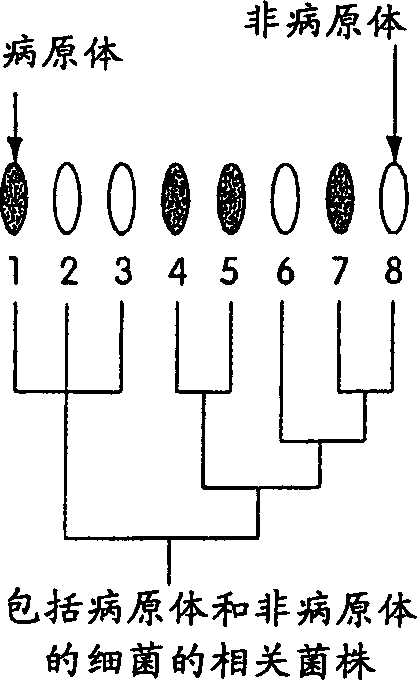
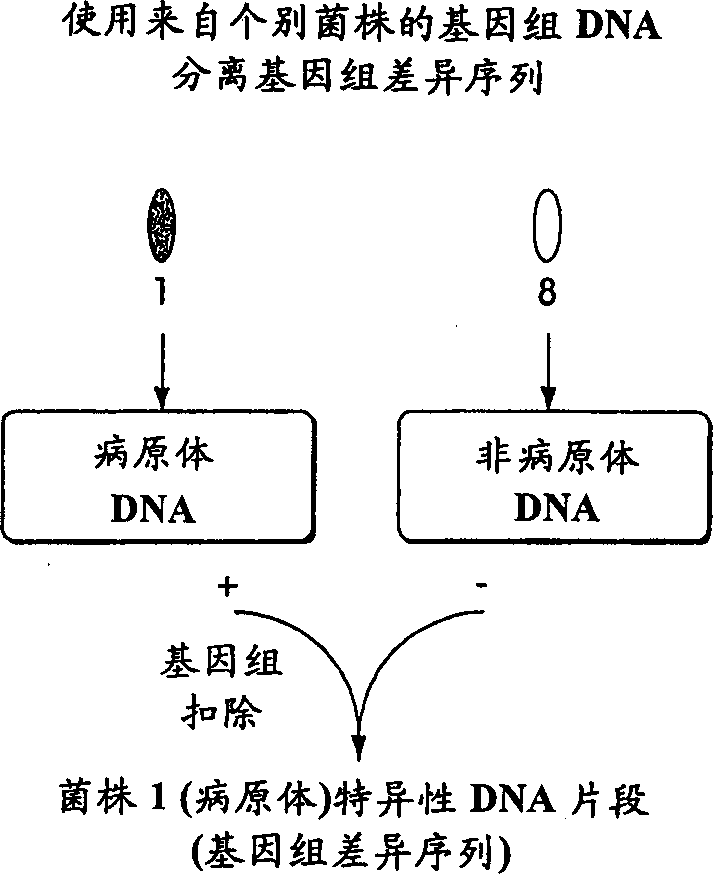
[0152] Step 1: Designate a collection of ID sequences including genomic difference sequences and group-specific sequences, which will be probed in a given test. This step involves selecting the organism to be tested and selecting a family of diagnostic ID sequences.
[0153] The first step in genomic profiling involves selecting the type of organisms to detect. For example, for medical applications, human pathogens can be selected; for detecting food spoilage, bacteria that cause food toxicity can be selected; for forensic purposes, multiple human individuals can be selected, and so on. Organisms selected for a particular test may vary widely in their genetic makeup, such as members of different kingdoms (i.e., viruses, bacteria, archaea, fungi, protozoa, plants, and animals); alternatively, the organisms selected may be a Smaller groups such as members of a species. An important application of genomic profilin...
Embodiment 2
[0319] Quantitative Analysis: What is the pathogen titer in the clinical sample? A powerful feature of genomic profiling assays is the ability to quantify pathogens in biological samples. Once target organisms have been identified by fingerprinting, their presence can be quantified by in situ hybridization with a portion of the original biological sample prepared according to standard methods (eg, Huang et al., Modern Pathology 11:971-977, 1998). I use a single molecule sensitive but simple method sufficient to detect nucleic acid sequences in a single organism (Huang et al., supra, 1998). This method is used with labeled probes for hybridization to the detection array. Alternatively, in situ hybridization can be performed using any of the biologically characteristic group-specific probes detectable by hybridization to the array. Example 2. Detection of the presence of pathogens in respiratory samples
[0320] pneumonia. Pneumonia is the most common cause of death from inf...
Embodiment 4
[0352] Identify pathogens present in blood samples. Blood samples were lysed and fingerprinted as described above for the reference strains, except ID probes from all bloodstream pathogen groups in Table 7 were included in the hybridization reaction. Pathogens present in the blood sample are identified by comparing the obtained fingerprints with those in a database of fingerprints of reference strains. Example 4. Forensic identification using genomic profiling assays
[0353] An overview of forensic identification. Identifying the origin of cellular samples is an important aspect of modern forensic analysis. Genetic identification of forensic samples requires amplification of DNA in cellular material, often only available in microscopic amounts, and comparison of the DNA with that of other individuals. Current methods of genetic identification generally require analytical gel electrophoresis, a step that is extremely time consuming and technically inappropriate for many for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com