Process for improving DNA detecting sensitivity
A technology of detection sensitivity and nano-metal particles, which is applied in the field of DNA detection, can solve the problems of DNA detection sensitivity decline and achieve the effect of increasing the degree of fixation and adsorption, and the same growth trend
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
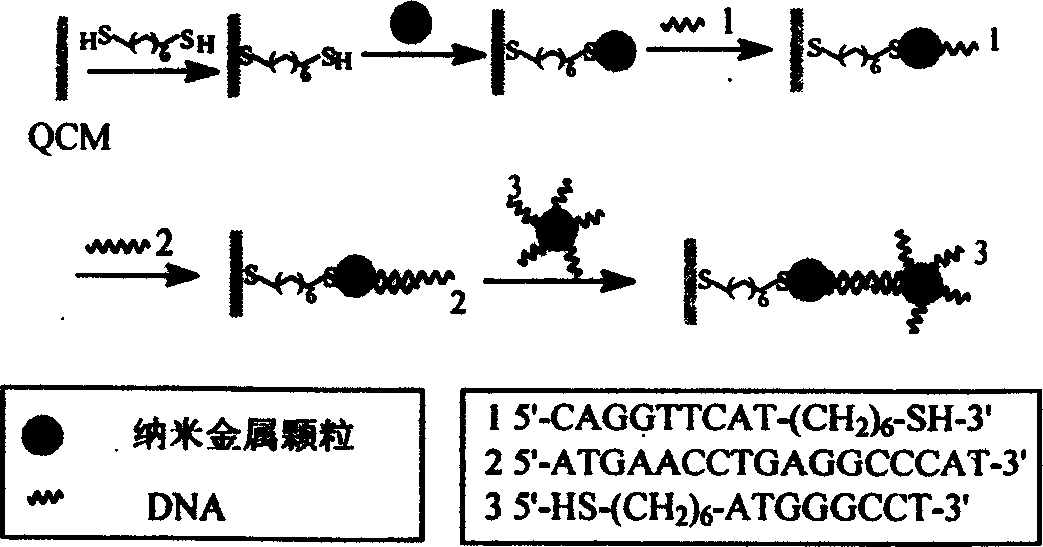
[0033] Example 1: 1,6-hexanedithiol was used as a linker to assemble 5 nm gold particles on the surface of QCM, and 20 nm gold particles were used as intensifying amplifiers for DNA detection
[0034] Take the AT-cut 9MHz QCM gold sheet, and use hot Piranha solution (98%H 2 SO 4 : 30%H 2 o 2 ) soaked for 3 minutes, fully rinsed with double distilled water, and measured its frequency with a quartz crystal microbalance. Modify the surface of QCM with 1,6-hexanedithiol, rinse with ethanol and twice double-distilled water after half an hour, and dry in the air. After QCM detection, add 20 microliters of 5nm gold sol dropwise, soak for 4 hours, rinse, and dry for measurement. frequency. Drop 2×10 -6 mol / L HS-DNA, after soaking for 1 hour, fully rinse and dry, then measure the frequency. Drop 2×10 -16 mol / L target DNA, after hybridization at 40°C for 2 hours, washed with phosphate buffer solution and twice redistilled water, and detected the frequency changes with a quartz cr...
Embodiment 2
[0035] Example 2: Use 1,6-hexanedithiol as linker to assemble 20nm gold particles on the surface of QCM and use 20nm gold particles as intensifying amplifiers for DNA detection
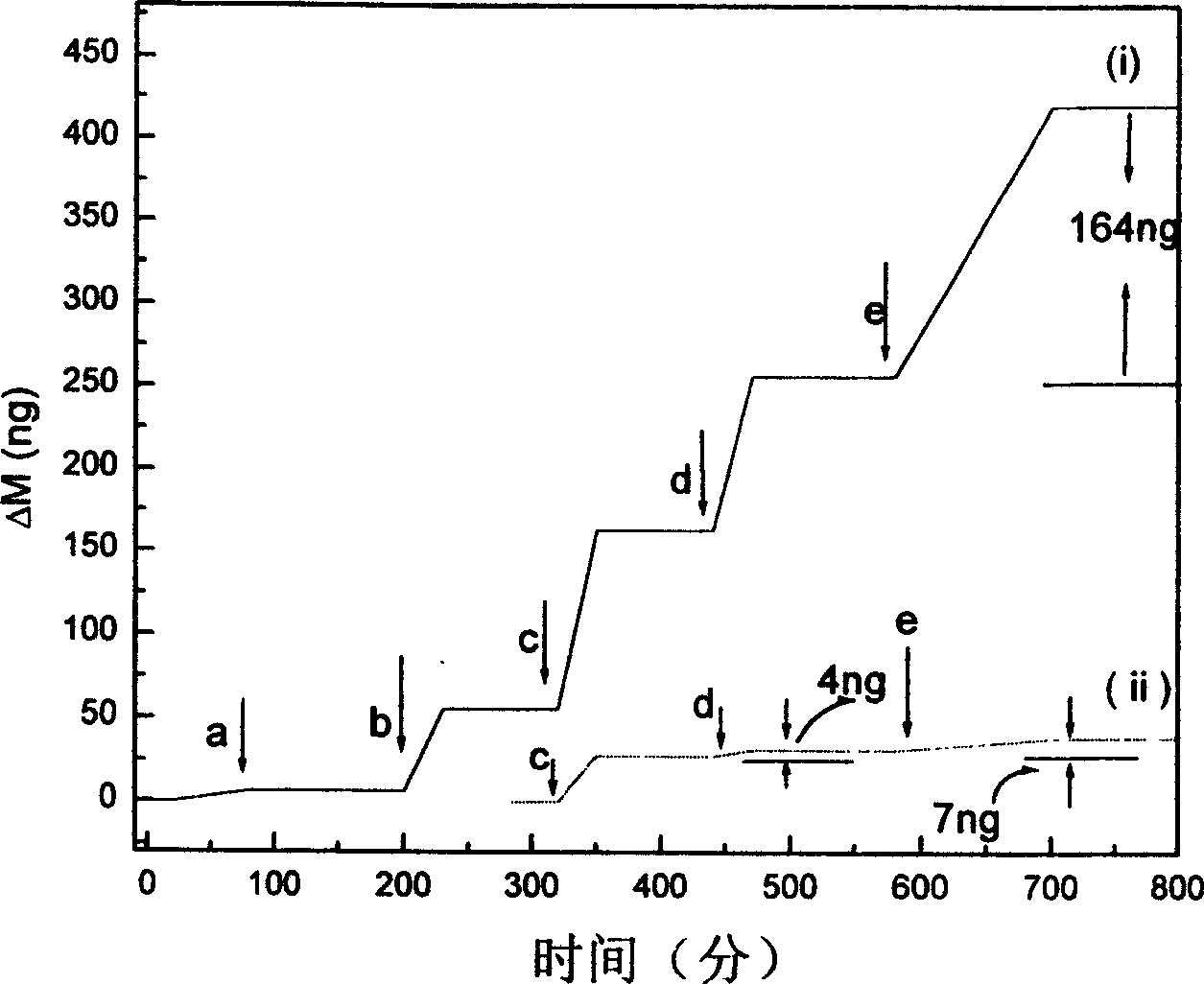
[0036] In a moist airtight chamber, drop 0.5% ethanol solution of 1,6-hexanedithiol onto the surface of QCM, rinse with ethanol and double-distilled water after half an hour, dry it, add dropwise after QCM detection 20 microliters of 20nm gold sol, soaked for 4 hours, rinsed, and dried for frequency measurement, there was a mass change of about 50ng. Drop 2×10 -6 mol / L HS-DNA, after soaking for 1 hour, fully rinsed and dried, and measured the frequency, the mass change was 107ng, while the mass change of HS-DNA immobilized directly on the bare QCM surface was 28ng. Drop 2×10 -16 mol / L target DNA, washed with phosphate buffer solution and double distilled water after hybridization at 40°C for 2 hours, the frequency change was 93ng. 20 microliters of DNA functionalized with 20nm gold sol was added dr...
Embodiment 3
[0038] Example 3: Using 1-amino 6-mercaptohexanethiol as linker to assemble 10nm gold particles on the surface of QCM and using 20nm gold particles as intensifying amplifiers for DNA detection
[0039] The surface of QCM was modified with 1-amino6-mercaptohexanol, and 20 microliters of 5nm gold sol was added dropwise after QCM detection, soaked for 4 hours, rinsed, and dried for frequency measurement. Drop 2×10 -6 mol / L HS-DNA, after soaking for 1 hour, fully rinse and dry, then measure the frequency. Drop 2×10 -16 mol / L target DNA, after hybridization at 40°C for 2 hours, washed with phosphate buffer solution and twice redistilled water, and detected the frequency changes with a quartz crystal microbalance. Hybridize with 20 microliters of DNA functionalized with 20nm gold sol, and use a quartz crystal microbalance to detect its frequency change and find that its detection sensitivity can reach 10 -16 mol / L.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com