Novel ligand detecting method
A detection method and a ligand technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as low sensitivity, poor accuracy, and complicated operation, and achieve high sensitivity, easy operation, and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
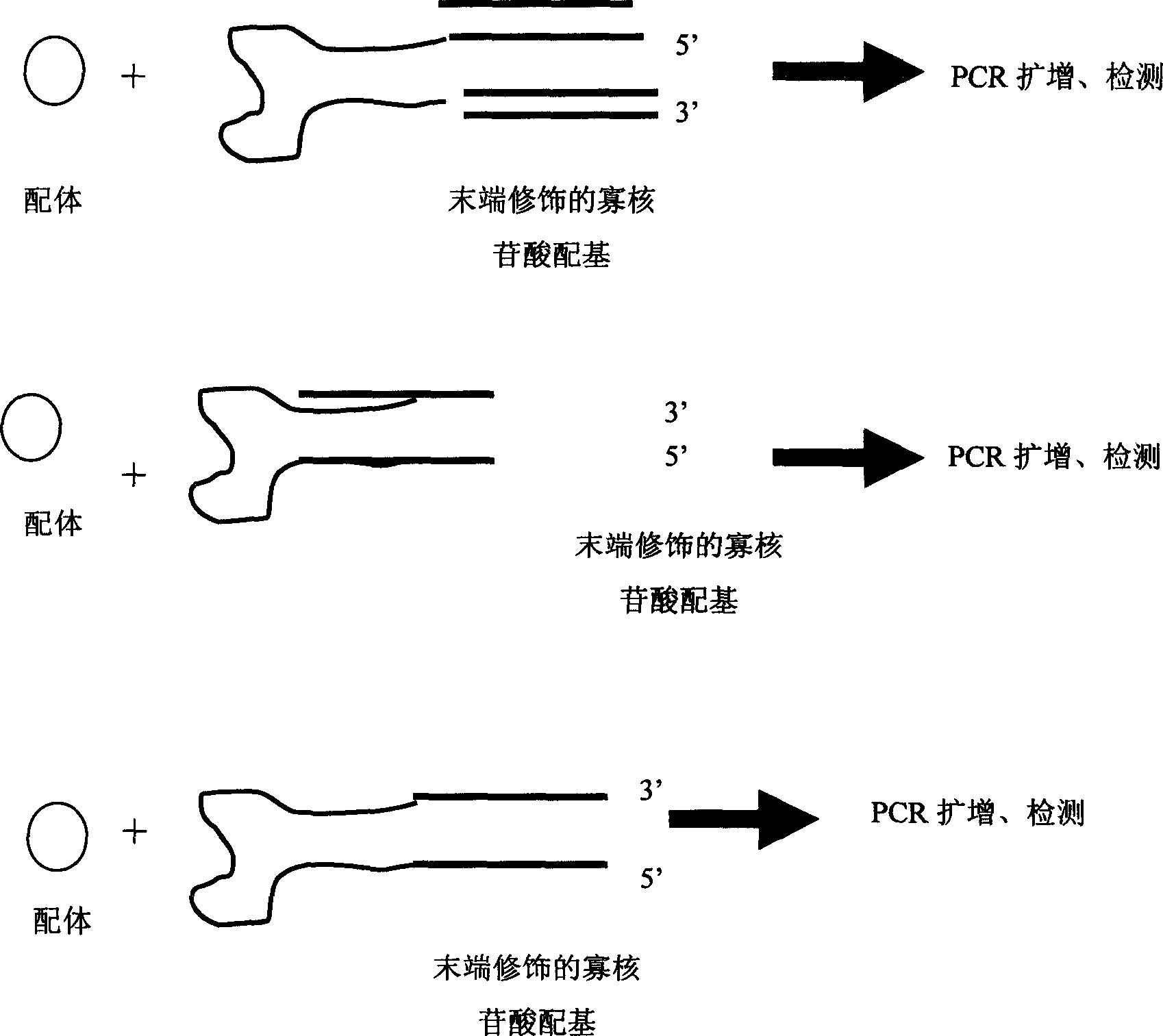
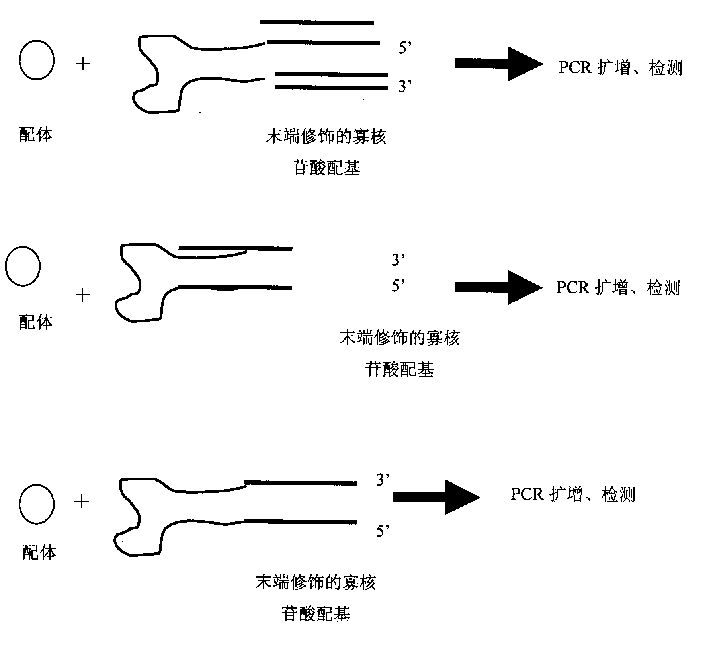
[0037] refer to figure 1 , using the ligand-specific oligonucleotide ligand modified by the nucleic acid sequence to directly bind the ligand, the ligand signal is converted into a nucleic acid signal, and then amplified by PCR to detect the ligand.
[0038] The principle and technical route of this embodiment are: fix the ligand to be tested on a medium such as a nitrocellulose membrane, seal the nitrocellulose membrane with a blocking solution, and then place the membrane with the ligand-specific oligonucleotide modified by nucleic acid sequence The acid ligand is incubated for a certain period of time, the cellulose membrane is fully washed with washing solution, the unbound specific oligonucleotide ligand is removed, and the ligand-specific oligonucleotide modified by the nucleic acid sequence combined with the ligand is amplified by PCR technology Acid ligand. The primers used in PCR amplification are labeled with fluorescent reagents or isotopes, etc., and the detection...
Embodiment 2
[0041] No need for any linking molecules, but the ligand-specific oligonucleotide ligands modified by chemiluminescent substances, enzymes, fluorescence and isotopes are directly combined with the ligands, and the ligand signals are converted into molecular signals, and then through the molecular signal Recognition, detection of ligands.
[0042] The principle and technical route of this embodiment are: fix the ligand to be tested on a medium such as a nitrocellulose membrane, seal the nitrocellulose membrane with a blocking solution, and then combine the membrane with a ligand-specific oligonucleotide modified by a signal molecule The acid ligand is incubated for a certain period of time, and the cellulose membrane is fully washed with washing solution to remove the unbound specific oligonucleotide ligand, and the detection of the ligand signal is realized by detecting the intensity of fluorescence or isotope. The present invention emphasizes that in the ligand detection meth...
Embodiment 3
[0044] After the oligonucleotide ligand of a certain type of ligand is artificially modified by the nucleic acid sequence (such as artificially adding some nucleic acid sequences that do not affect its binding activity), according to the length of the modified sequence, the base sequence is different, through the oligonucleotide The specific combination of the nucleotide ligand and the ligand makes the modified sequence represent different ligands, and then amplified by PCR (or rolling circle replication) to detect the modified sequence, thereby identifying the ligand. This technology can be used to build microarrays or biochips.
[0045]The principle and technical route of this embodiment are: fix the test samples containing various ligands to be tested on media such as nitrocellulose membranes, seal the nitrocellulose membranes with blocking solution, and then combine the membranes with those modified with different nucleic acid sequences. After incubating the specific oligo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com