Method for identification of proteins from intracellular bacteria
A protein and bacteria technology, applied in the field of identifying intracellular bacterial proteins, can solve problems such as reducing tryptophan synthesis ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0235] Infection of mammalian cell cultures
[0236] According to the methods described in [19.] and [17.], use Chlamydia pneumoniae VR1310, Chlamydia trachomatis serotype A (HAR-13), D (UW-3 / Cx) or L2. (434 / Bu) (ATCC) ) An inclusion body forming unit (IFU) infected semi-confluent HeLa, HEp-2 or McCoy (ATCC, Rockville, MD, USA) cell monolayer. The infection medium for Chlamydia trachomatis A and D consists of RPMI 1640, 25mM HEPES, 10% FCS, 1% w / v glutamine, and 10mg / ml gentamicin. The infection medium for Chlamydia trachomatis L2 consists of RPMI 1640, 25mM HEPES, 5% FCS, 1% w / v glutamine, 10mg / ml gentamicin.
Embodiment 2
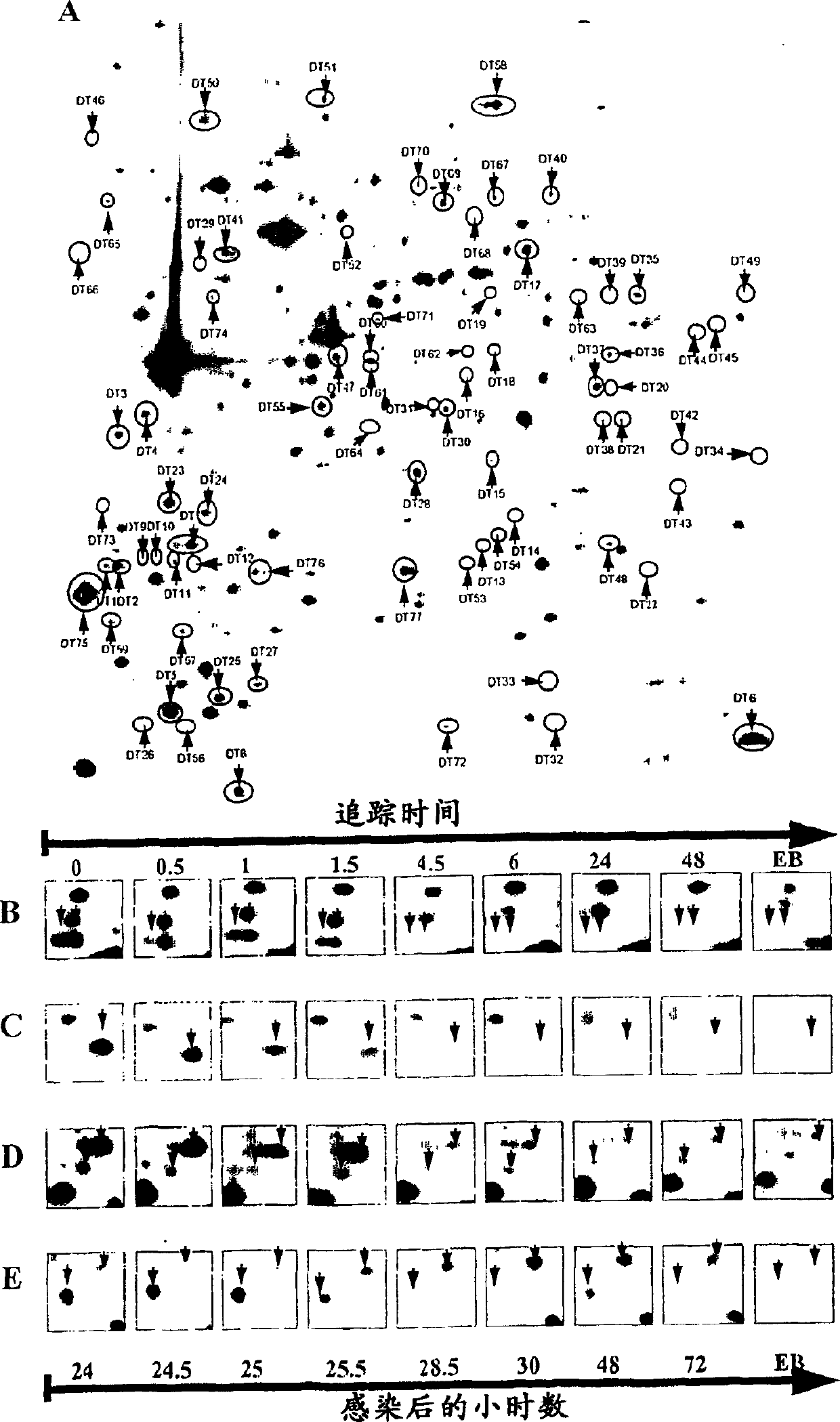
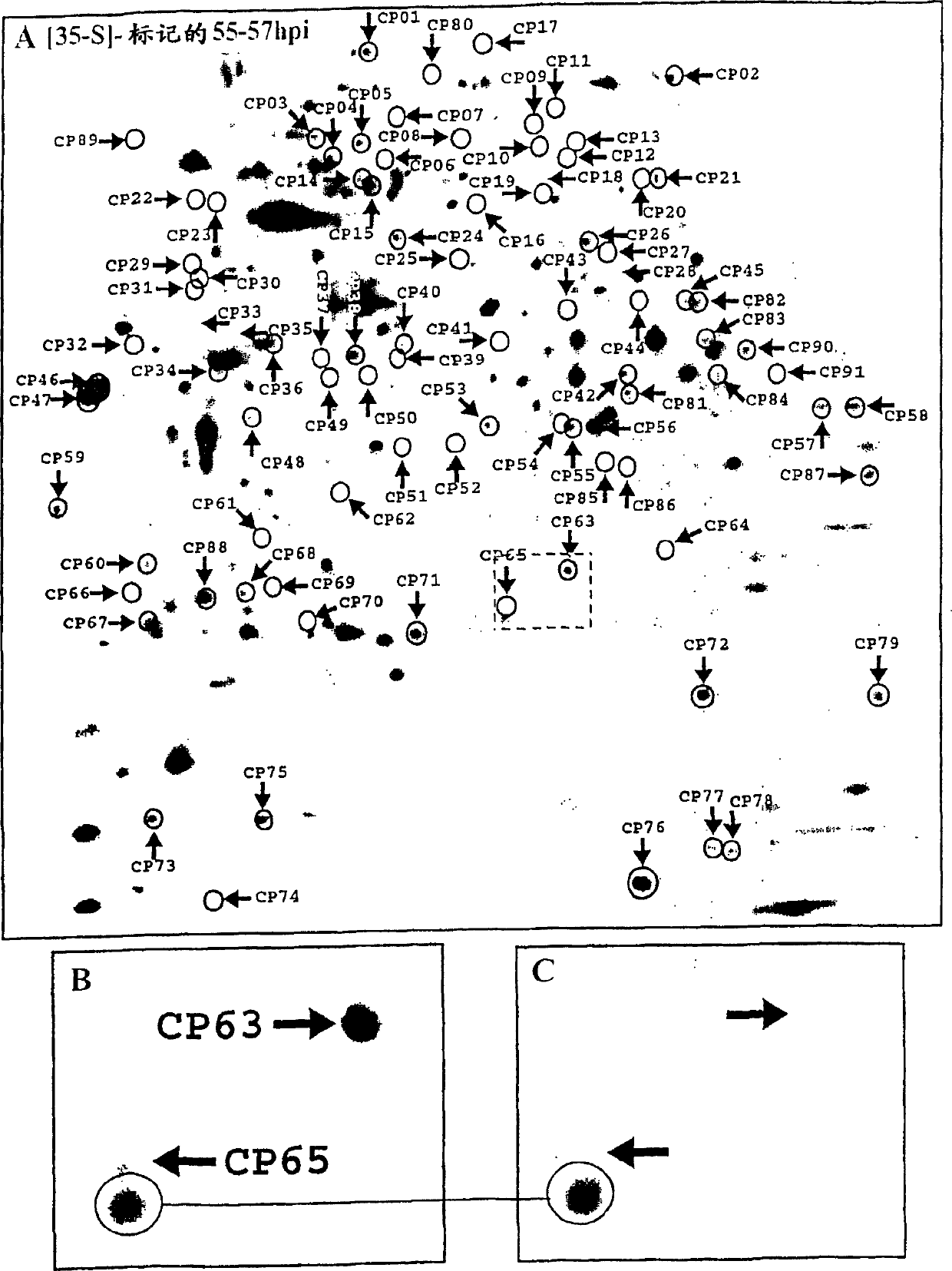
[0238] Pulse marking / tracking analysis
[0239] In order to label the chlamydia protein for a period of two hours, according to the above-mentioned method (Shaw et al., 1999, 2000) [18.] [19.], containing RPMI 1640, 10 mg / ml gentamicin, 40 μg / ml actinomycetes The infected cells were cultured in a ketone, 100 μCi / ml [35S]-methionine / cysteine (Promix, Amersham Pharmacia Biotech, Uppsala, Sweden) medium. After labeling, the labeling medium was replaced with a normal medium by washing twice with normal medium, and the infected cells were collected at different time points after the labeling. Similarly, the labeled EB protein was obtained by culturing the chlamydia for two hours after labeling to 72 hours after infection. The labeled EBs are then collected and purified using two consecutive steps of density gradient ultracentrifugation, which is basically as used for Chlamydia trachomatis (Schacter and Wyrick, 1994) [22.] and pneumonia Chlamydia (Knudsen et al. 1999 [17.]) density g...
Embodiment 3
[0241] Sample preparation
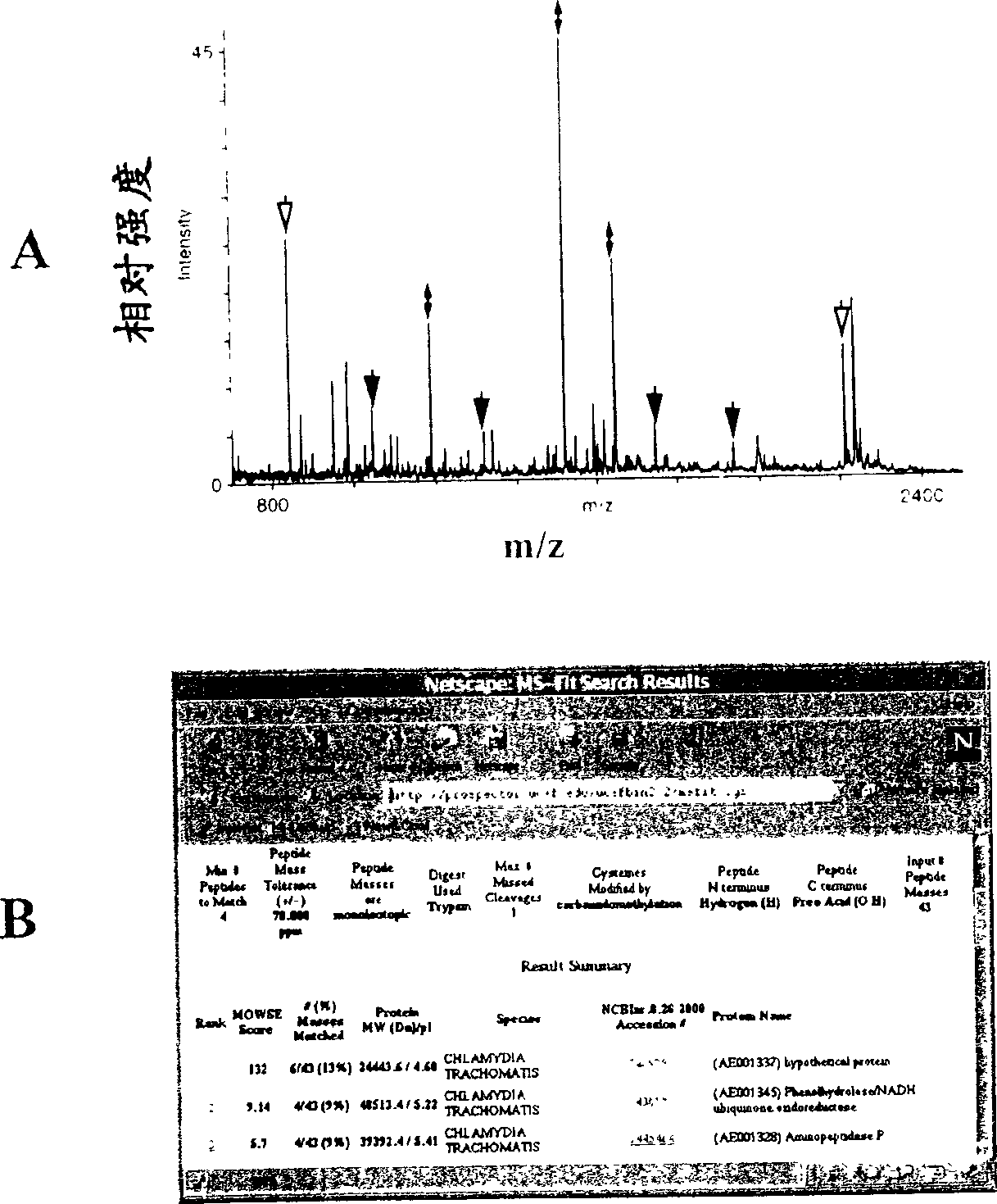
[0242]After [35S]-labeled, the cells were washed twice with PBS and then dissolved in a standard lysis buffer containing 9M urea, 4% w / v 3-[(3-cholamidopropyl)dimethyl Ammonium]-1-propanesulfonate (CHAPS; Roche, Germany), 40 mM Tris Base, 65 mM DTE, and Pharmalyte 3-10 (Amersham Pharmacia Biotech). In order to enrich high molecular weight hydrophobic proteins, 7M urea, 2M thiourea, 4% w / v 3-[(3-cholamidopropyl)dimethylammonium]-1-propanesulfonate (CHAPS ; Boehringer Mannheim, Germany), 40mM Tris Base, 65mM dithioerythritol (DTE) and 2% v / vPharmalyte 3-10 (Amersham Pharmacia Biotech), basically in accordance with (Harder et al. 1999 [23.]) The proceeding. Sonicate the sample containing intact fine lysate or purified EB, and centrifuge at 10000×g for 10 minutes. Store the sample at -70°C until use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com