Process for testing bacteria gene chip in sewage
A kind of technology of gene chip and detection method, which is applied in the field of gene chip detection of bacteria in sewage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
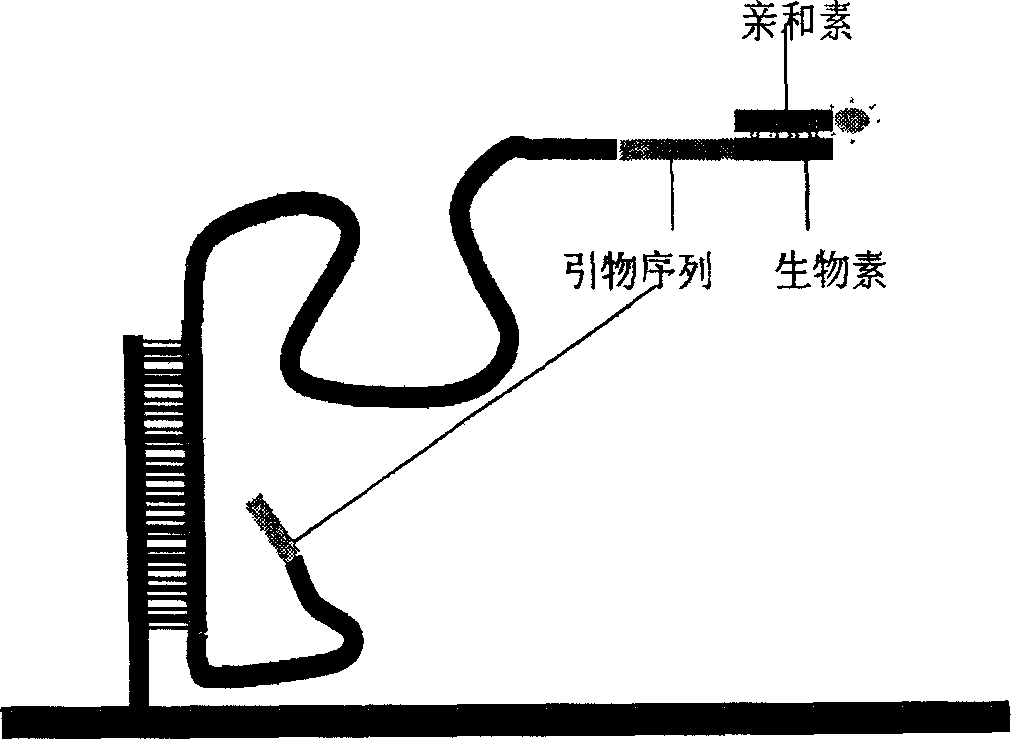
Image
Examples
Embodiment 1
[0035] Embodiment 1: In this embodiment, enterococcus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Shigella, Salmonella, Escherichia coli 0-157 and Escherichia coli are selected as the target bacteria in the food processing sewage water sample .
[0036] 1. Specific design of bacteria-specific primers and species and genus detection probes:
[0037] First login to GeneBank ( http: / / ncbi.nlm.nih.gov / entrez ), searched in the nucleic acid database, and obtained the 16SrRNA gene sequences of the above-mentioned 7 kinds of bacteria. The ClustalW Alignment program of the Mac Vector6.5.1 software package was used for sequence alignment, and the base alignment graph and cluster analysis results were obtained. The constant and variable regions of the 16S rRNA gene were divided according to the base alignment diagram. Use Primer5.0 software to design double-stranded primers in the variable region, compare the obtained candidate double-stranded primers with BLAST, screen out the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com