Important controlling gene PIX in anti-tumor immunological reaction
A technology of immune regulation and NK cells, applied in the field of cell biology, can solve problems such as defects in NK cell signaling pathways
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Yeast two-hybrid screening and verification
[0045] According to the known SAP gene sequence (GenBank accession number: AL023657), using the lymphocyte cDNA library as a template, using primers 5'ATGGACGCAGTGGCTGTGTATC (SEQ ID NO: 1), 3'TCATGGGGCTTTCAGGCAGAC (SEQ ID NO: 2), by PCR technology Obtain the full length of the SAP gene, connect it to the vector pGB (Clonetech Company) containing the BD functional domain (binding domain) to obtain the plasmid pGB-SAP as bait, and screen the B lymphocytes through the yeast two-hybrid system (Clonetech: yeast-two-hybridsystem2) Cell and T lymphocyte cDNA library, the obtained clones were sequenced and identified as multiple clones of SLAM, 2B4, α-PIX, β-PIX (P21 protein-activated kinase-associated exchange factor). The screening results of the yeast two-hybrid system are shown in Table 1.
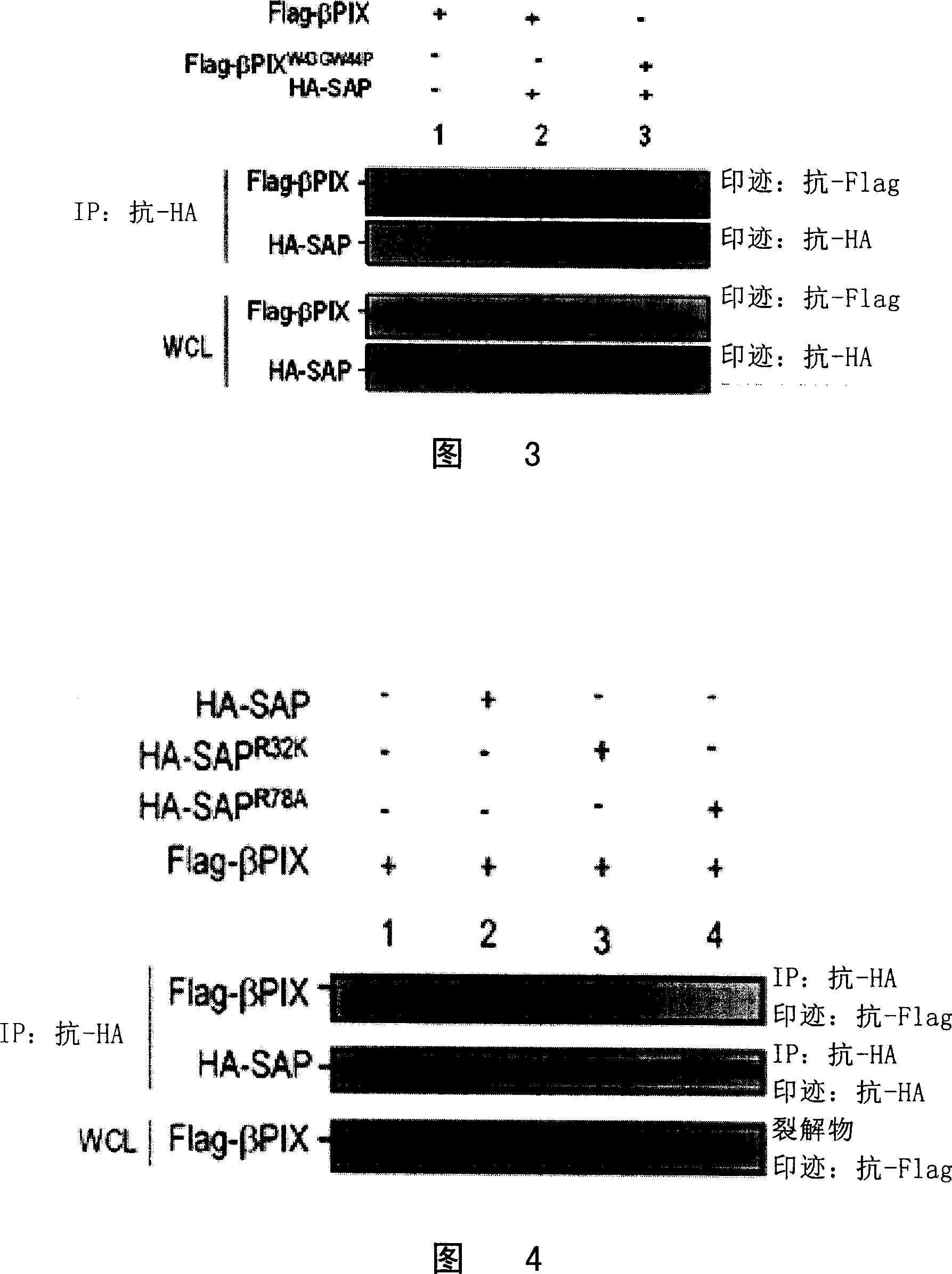
[0046] Using the above pGB-SAP wild type as a template, use the point mutation construction kit QuikChangeSite-Directed Mutagene...
Embodiment 2
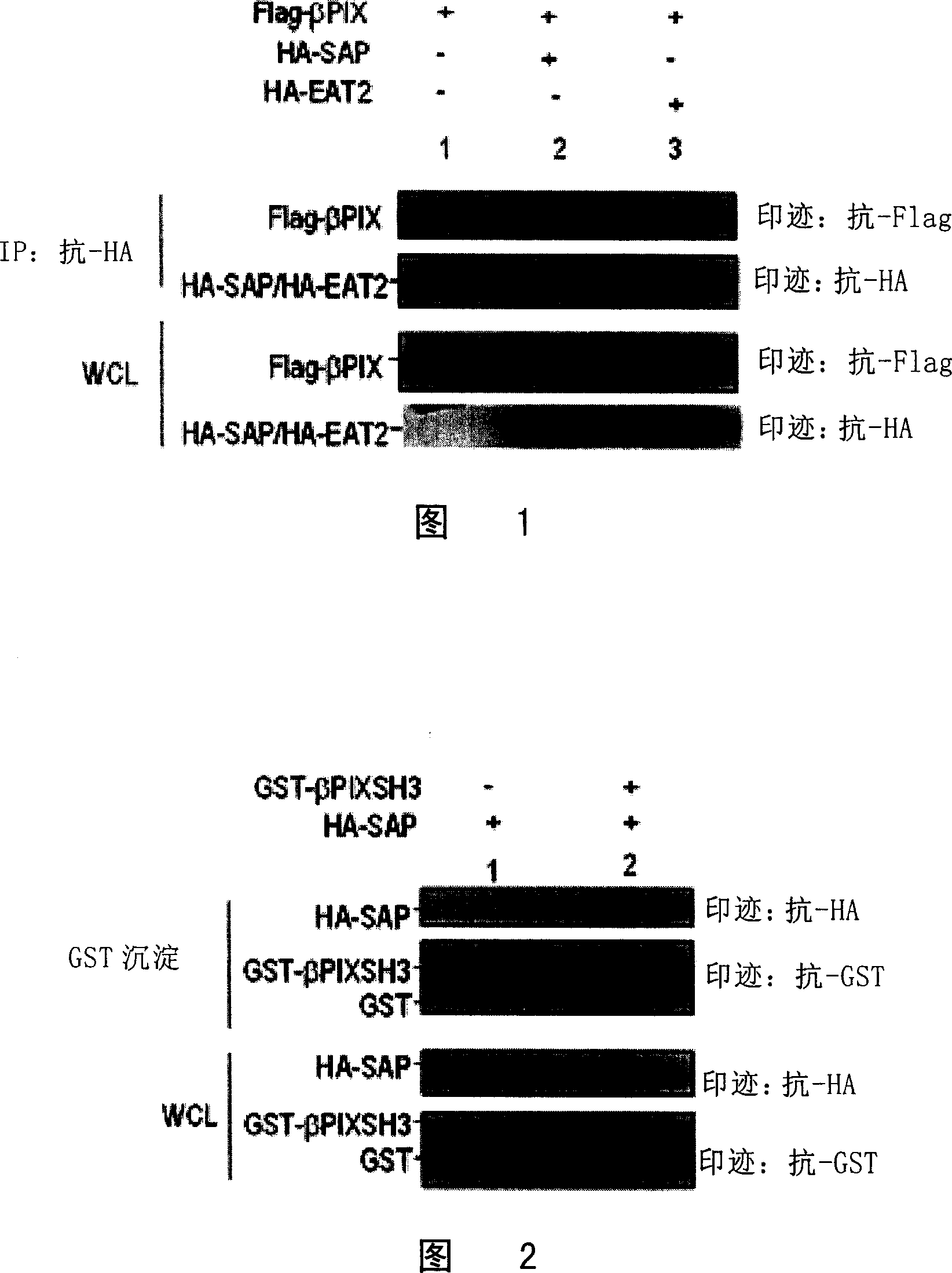
[0051] Example 2 Verifies the interaction between SAP and β-PIX in mammalian cells
[0052] 1. In mammalian cells, SAP, EAT2 interact with β-PIX
[0053]The SAP and β-PIX gene fragments in the above yeast plasmids pGB-SAP and pACT2-β-PIX were cloned into the eukaryotic expression vector pCDEF containing Flag and HA tag (see US Patent US20020076415A1) to form pCDEF-HA-SAP and pCDEF-Flag-β-PIX. At the same time, according to the known EAT2 gene sequence (GenBank accession number: AF256653), using the lymphocyte cDNA library as a template, using primers 5'ATGGATCTGCCTTACTACCATGGAC (SEQ ID NO: 7), 3'TCAAGGCAAGACATCCACATAATCGC (SEQ ID NO: 8), by PCR Technology Obtain the full length of the EAT2 (SAP analogue in B lymphocytes) gene and clone it into the pCDEF-HA vector to obtain the pCDEF-HA-EAT2 plasmid. The obtained plasmid was co-transfected into mammalian cell 293T (ATCC). After 24 hours of culture, the cells were broken, and the supernatant of the lysate was collected, and wh...
Embodiment 3
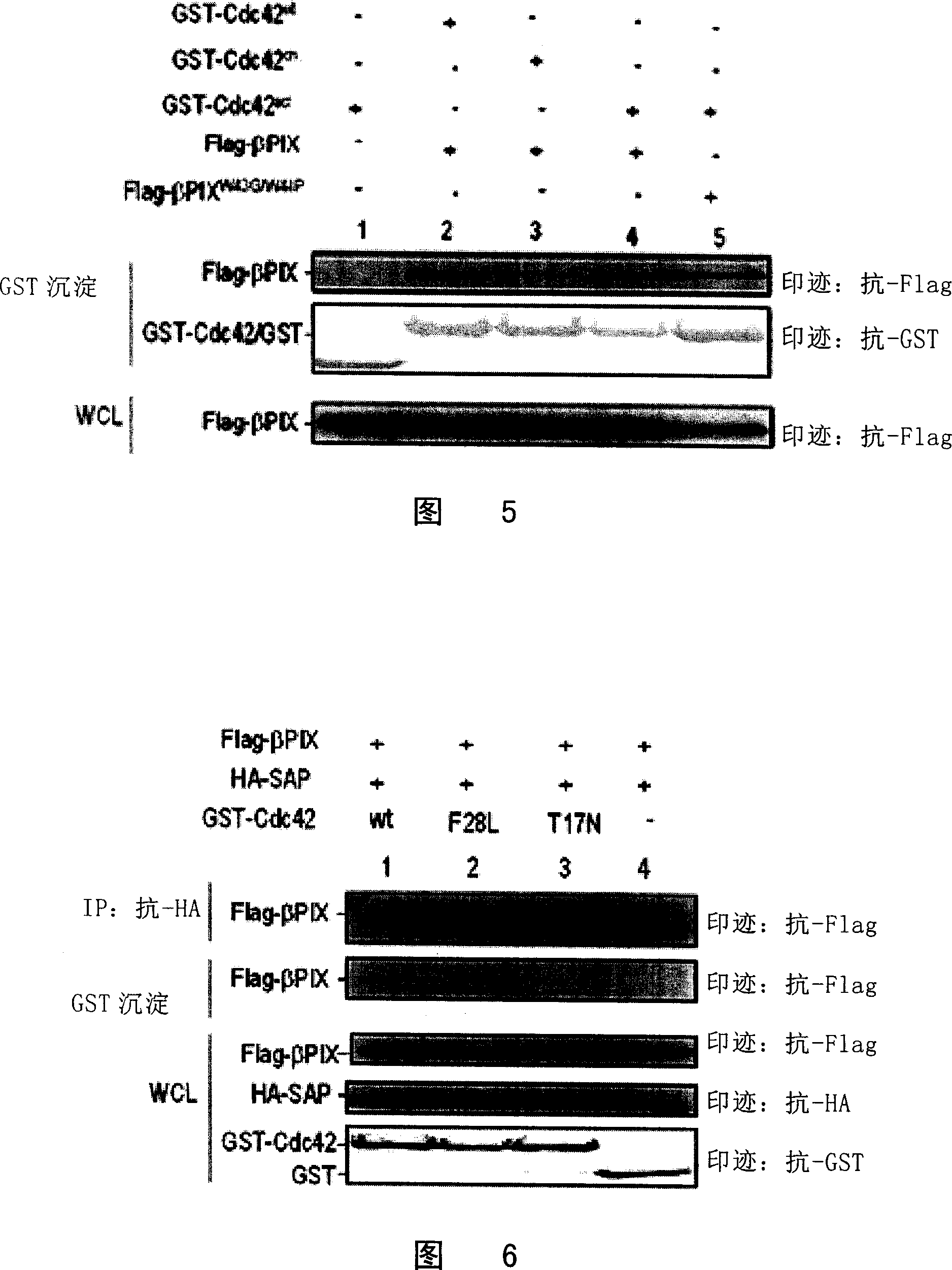
[0067] Example 3 Study on the interaction of SAP with β-PIX and Cdc42 by co-immunoprecipitation technique
[0068] 1. Validate the interaction between β-PIX and Cdc42 by co-immunoprecipitation technique
[0069] According to the known Cdc42 (Homo sapiens cell division cycle 42) gene sequence (GenBank accession number: NM_001791), using the lymphocyte cDNA library as a template, primers 5'ATGCAGACAATTAAGTGTGTTGTTGTG (SEQ ID NO: 9); 3' TCATAGCAGCAGCACACACCTGCGG (SEQ ID NO: 10), the full length of the Cdc42 gene was obtained by PCR technology, and connected to the pCDEF-GST vector to obtain pCDEF-GST-Cdc42. Using this as a template, the Cdc42 mutant pCDEF-GST-Cdc42 in the activated state was constructed by the point mutation method (constructed with the SAP point mutation in Example 1) F28L和 Inactive Cdc42 mutant pCDEF-GST-Cdc42 T17N . The pCDEF-GST empty vector, pCDEF-GST-Cdc42, pCDEF-GST-Cdc42 F28L , or pCDEF-GST-Cdc42 T17NCo-transfect mammalian cells 293T with pCDEF-Flag-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com