Predicting breast cancer treatment outcome
A breast cancer and anti-breast cancer technology, applied in the direction of biomass pretreatment, microbial measurement/inspection, biomass post-treatment, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0084] In a first aspect, the present invention relates to gene expression patterns (or profiles or "signatures") capable of distinguishing (or correlating) the survival of breast cancer patients treated with tamoxifen (TAM) or other "anti-estrogen" drugs against breast cancer. ”) identification and application. The methods of the invention employ a number of control cells or tissue samples, such as those known to those of ordinary skill in the art of breast cancer pathology, to be breast cancer cells rather than normal or other non-cancerous cells to identify such expression patterns. Patient outcomes can be correlated with expression data to identify expression patterns that correlate with outcomes following treatment with TAM or other anti-breast cancer "anti-estrogen" drugs. Because the overall gene expression profile varies from person to person, cancer to cancer, and cancer cell to cancer cell, certain cells can be correlated with expressed or underexpressed genes as des...
Embodiment
[0440] Example
[0441] general method
[0442] Patient and tumor selection criteria and trial design
[0443] The criteria for the patients in this study were: female patients with ER-positive breast cancer diagnosed at Massachusetts General Hospital (MGH) during 1987-2000, treated with standard breast surgery (modified radical mastectomy or lumpectomy) ) and radiotherapy followed by 5 years of systemic adjuvant tamoxifen; the patient had not received chemotherapy prior to relapse. Clinical and follow-up data were obtained from the MGH Tumor Registry. There were no missing enrollment data and secondary data validation was performed on all available medical records.
[0444] All tumor specimens collected at initial diagnosis were obtained from frozen and formalin-fixed paraffin-embedded (FFPE) tissue banks at Massachusetts General Hospital. Tumor samples with more than 20% tumor cells were selected, and the median of all...
Embodiment 2
[0467] Identification of differentially expressed genes
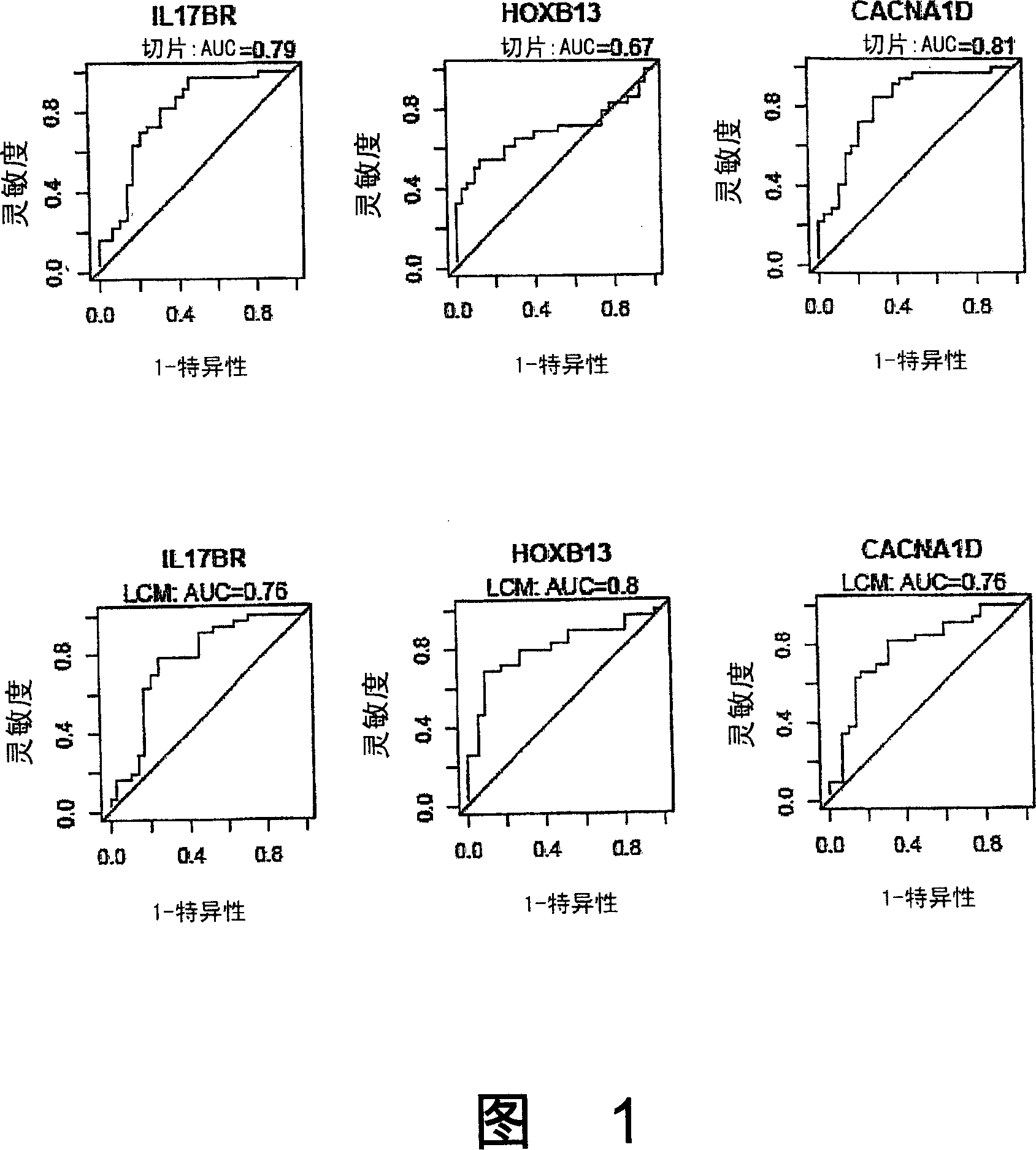
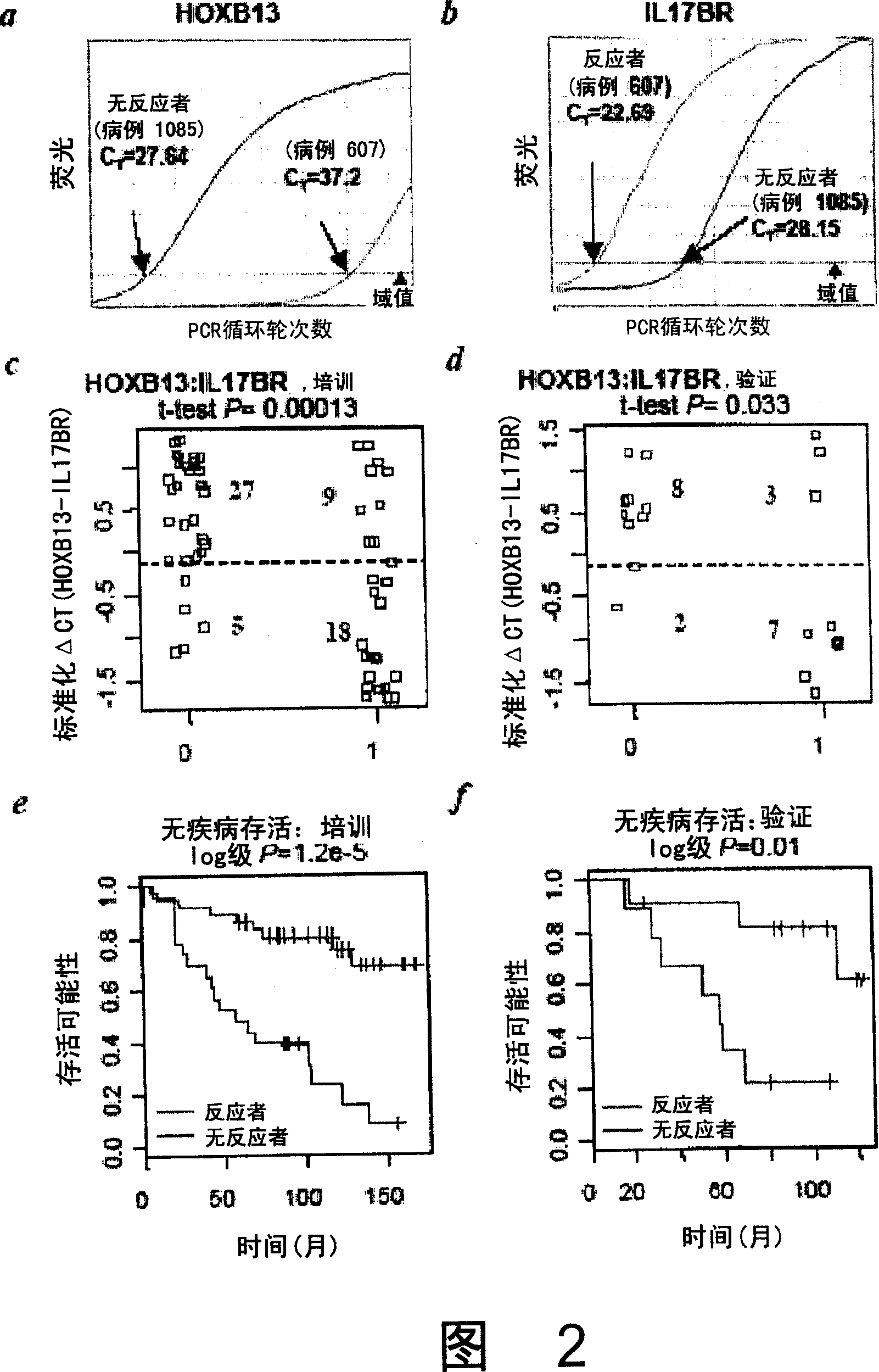
[0468] Gene expression pattern analysis was performed using the 22,000-gene oligonucleotide microarray described above. In the initial analysis, RNA isolated from frozen tumor tissue sections taken from the primary biopsy was used. The resulting expression data set was first filtered according to the total variance of individual genes, and the top 5,475 highly differential genes (75%) were selected for further study. Using this reduced data set, a t-test comparing tamoxifen responders and lung responders was performed for each gene, resulting in the identification of 19 differentially expressed genes with a P value of 0.001 (Table 2). For treatment outcomes random permutation of patient types and repetition of the t-test procedure 2,000 times, the estimated probability of selecting by chance such a much differentially or more differentially expressed gene is about 0.04. Thus, this analysis reveal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com