Novel oligonucleotide arrays and their use for sorting, isolating, sequencing, and manipulating nucleic acids
a technology of oligonucleotide arrays and arrays, which is applied in the field of sorting, isolating, sequencing, and manipulating nucleic acids, can solve the problems of inability to unambiguously determine inability to identify inability to detect the sequence of dna, etc., to achieve increase the specificity of hybridization, and increase the effect of hybridization specifi
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0043] Throughout the detailed description, references to the examples section are made to illustrate particular embodiments of the aspect of the invention discussed. Also, techniques described with respect to one embodiment may not be explicitly described in other embodiments. Their application to the several embodiments described herein, however, is understood.
[0044] All periodicals, patents and other references cited herein are hereby incorporated by reference.
I. OLIGONUCLEOTIDE ARRAYS
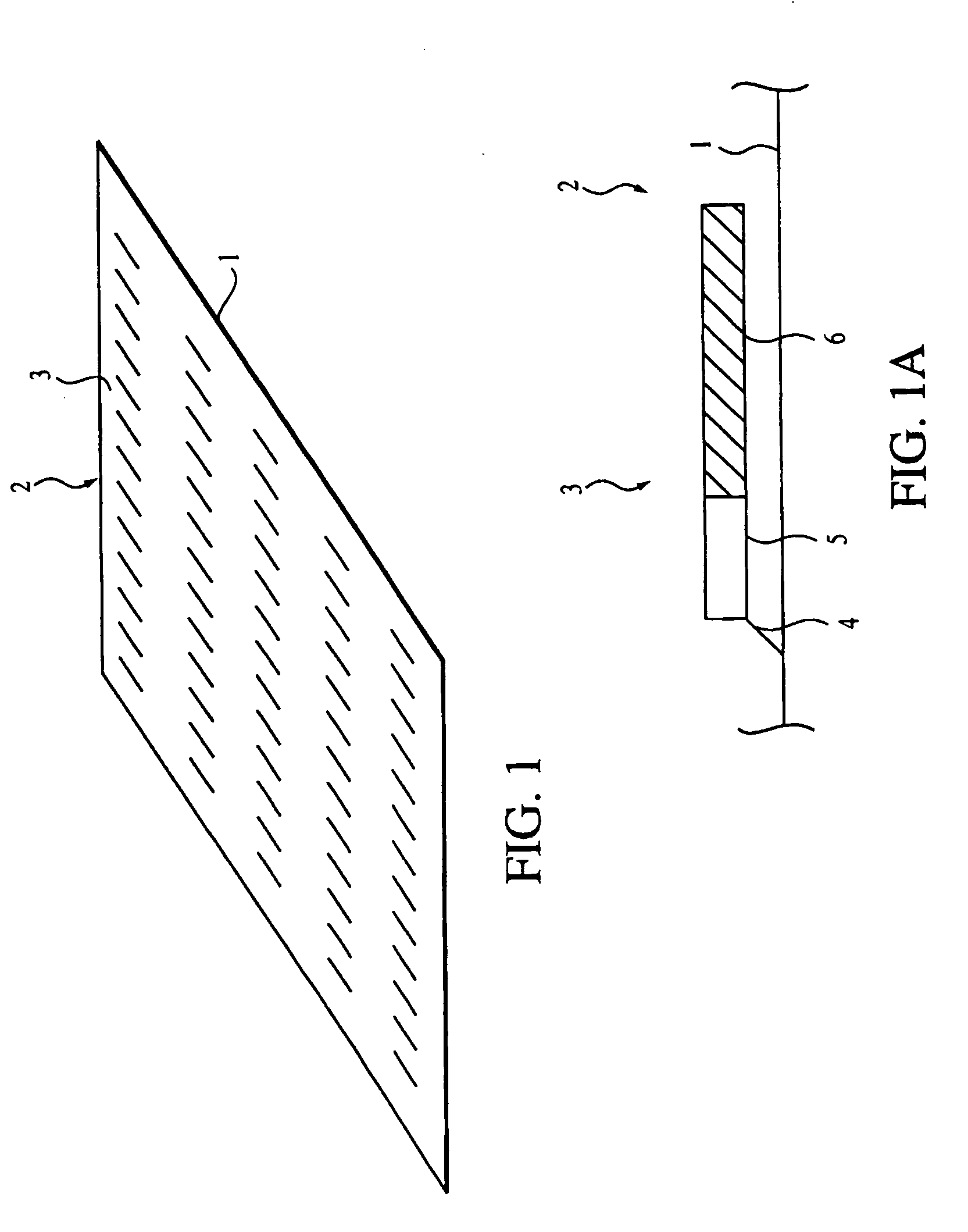
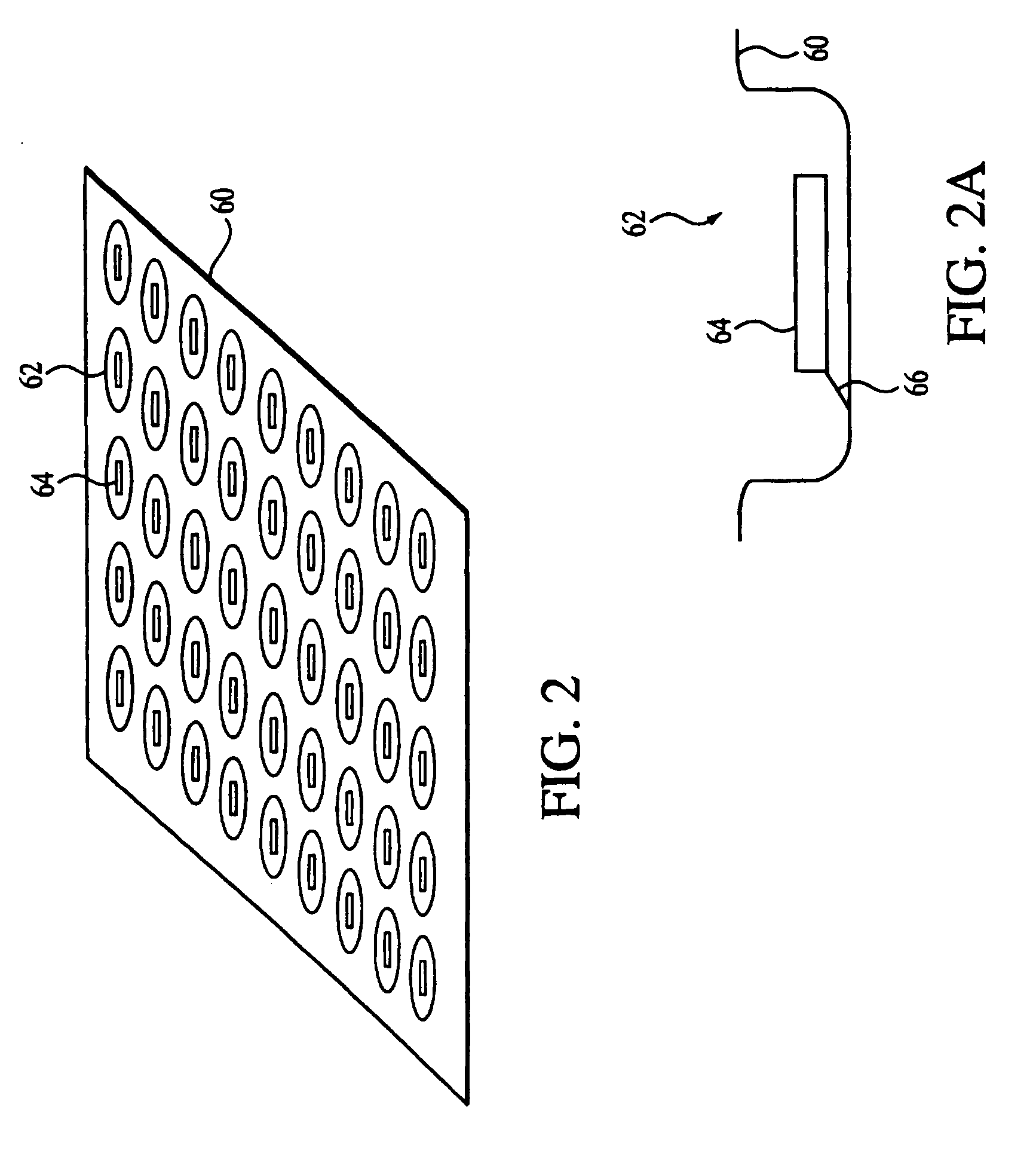
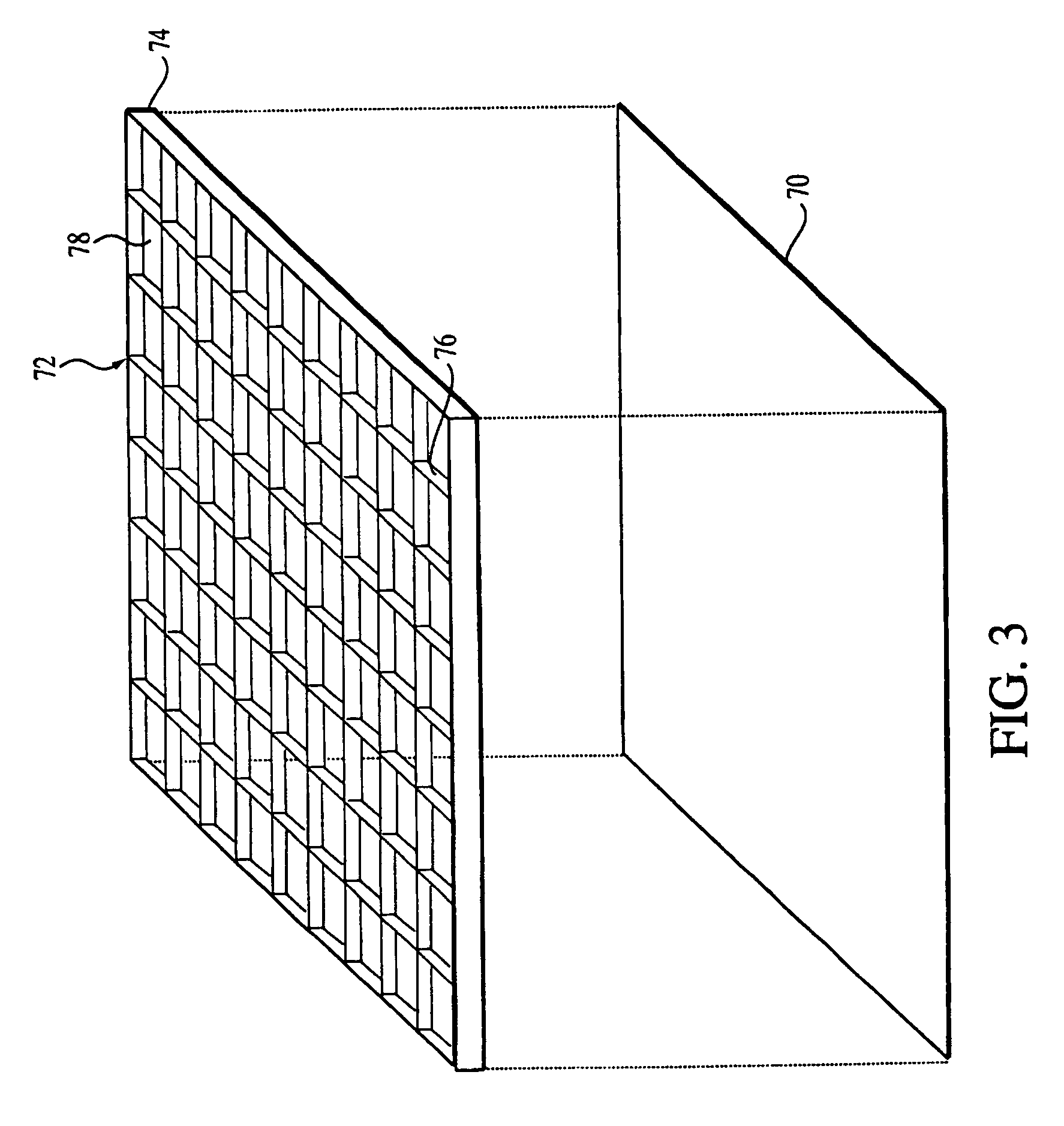
[0045] As used herein an “oligonucleotide array” is an array of regularly situated areas on a solid support wherein different oligonucleotides are immobilized, typically by covalent linkage. Each area contains a different oligonucleotide, and the location within the array of each oligonucleotide is predetermined. If the array is made of oligodeoxyribonucleotides, the nucleotides are: deoxyadenylate (dA), deoxycytidylate (dC), deoxyguanylate (dG), and deoxythymidylate (dT) (for brevity, the prefix...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com