Polynucleotides and methods for making plants resistant to fungal pathogens
a technology of fungal pathogens and polynucleotides, which is applied in the field of polynucleotides and methods for making plants resistant to fungal pathogens, can solve the problems of increasing problems, affecting the harvesting of lodged plants, and affecting the quality of the finished produ
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
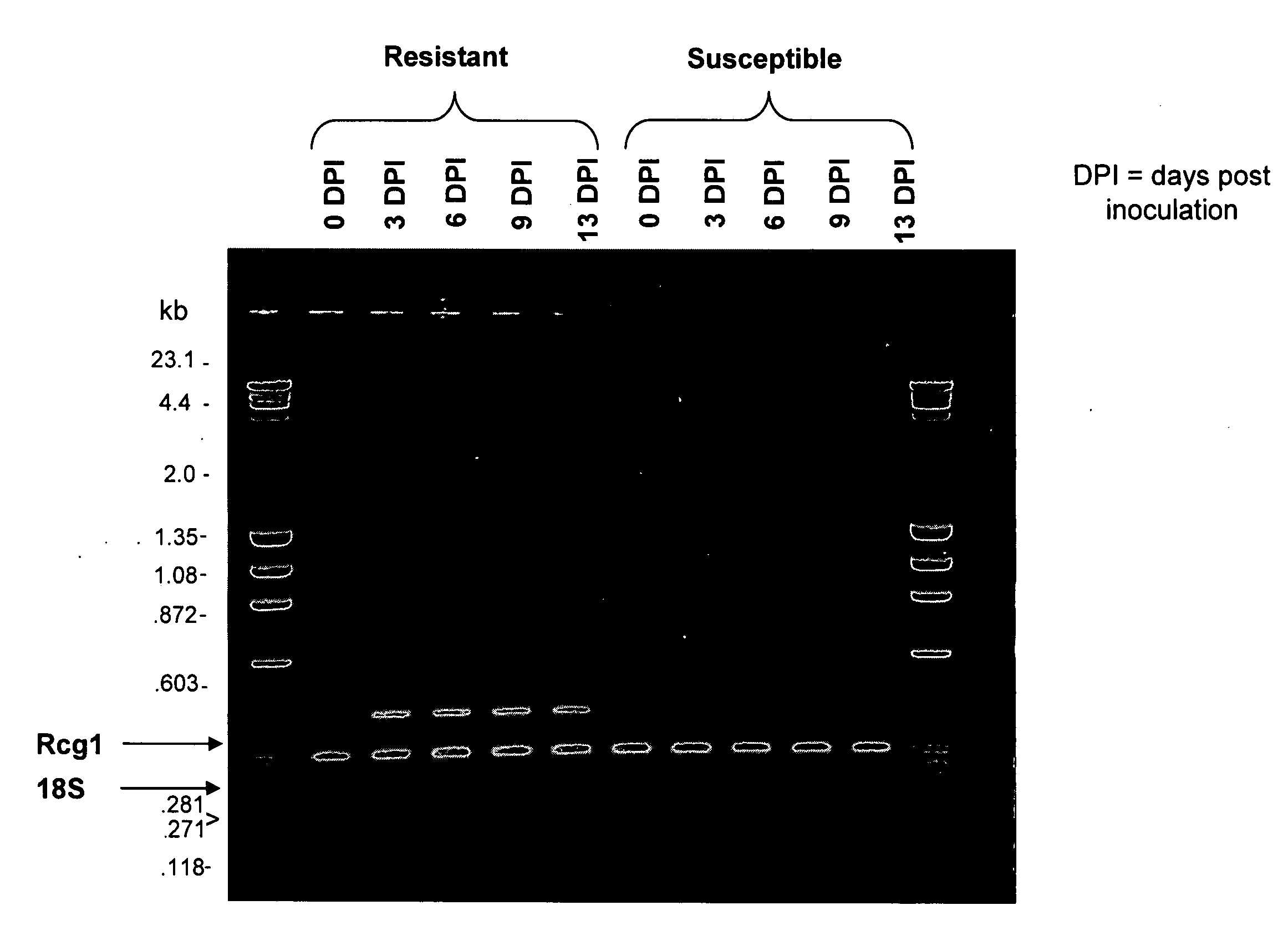
Fine Mapping of the Rcg1 Locus to a Specific Region of 4L
[0185] In order to map and clone the gene responsible for the resistance of corn line MP305 to Cg, lines had previously been created which differed as little as possible from each other genetically with the exception of the presence of the locus responsible for the resistant phenotype. Such lines are called near isogenic lines. To this end, DE811 had been crossed to MP305 and the progeny had been backcrossed to the sensitive line DE811 three times, at each backcross selecting for resistance to Cg and otherwise for characteristics of DE811 (Weldekidan and Hawk, (1993), Maydica, 38:189-192). The resulting line was designated DE811ASR (BC3) (Weldekidan and Hawk, (1993) supra). This line was used as the starting point for the fine mapping of the Rcg1 locus. It was first necessary to know roughly where in the maize genome it was located. Using standard genetic methods, Jung et al. ((1994) supra) had previously localized the locus ...
example 2
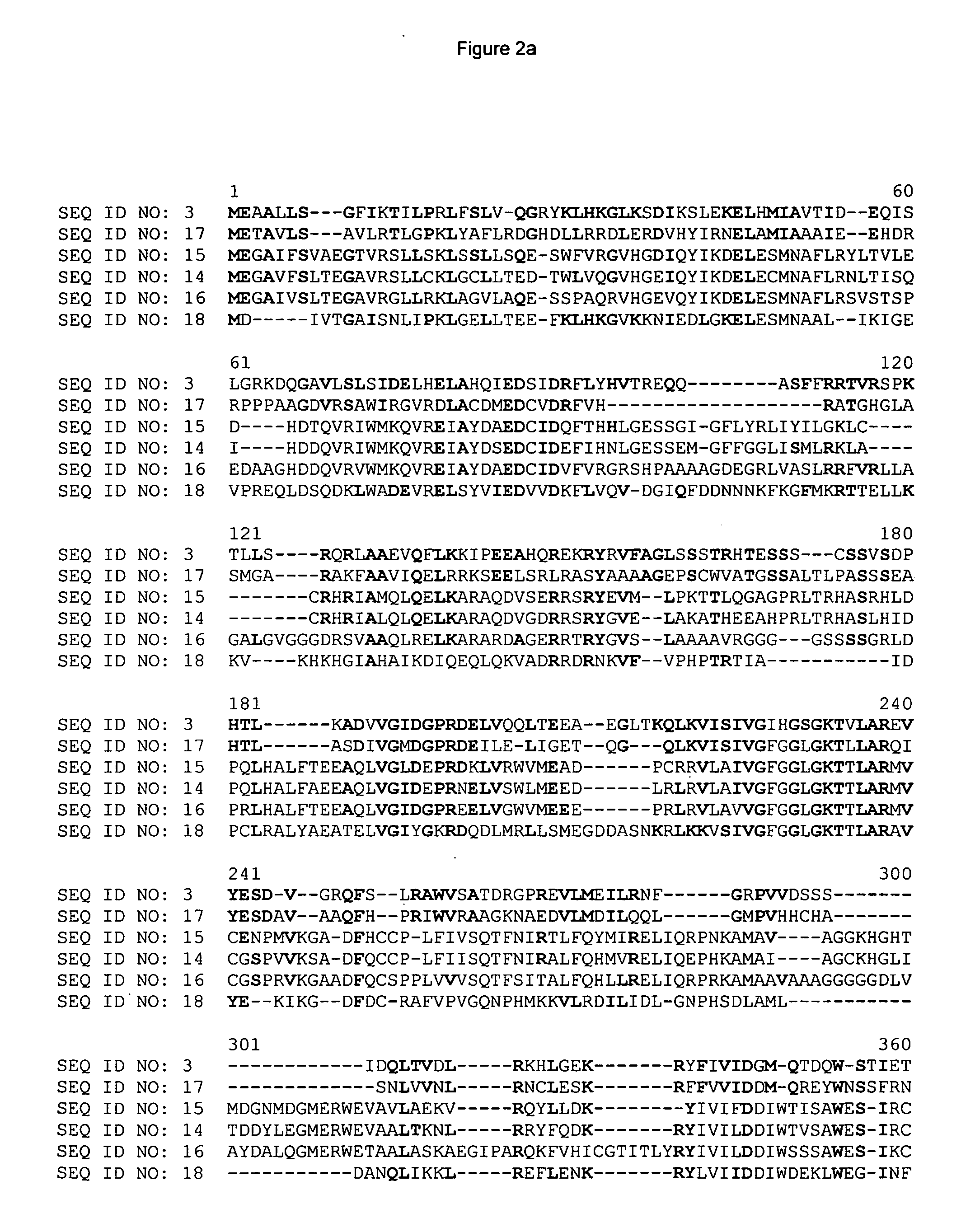
Isolation of BAC Clones from the Resistant Lines and Identification of Candidate Genes in the Region of the Rcg1 Locus
[0193] In order to isolate the gene responsible for the phenotype conferred by the Rcg1 locus, BACs containing the region between the FLP 8 and FLP 27 markers were isolated from a BAC library prepared from the resistant line DE811ASR (BC5). This library was prepared using standard techniques for the preparation of genomic DNA (Zhang et al. (1995) Plant Journal 7:175-184) followed by partial digestion with HindIII and ligation of size selected fragments into a modified form of the commercially available vector pCC1 BAC™ (Epicentre, Madison, USA). After transformation into EPI300™ E. coli cells following the vendors instructions (Epicentre, Madison, USA), 125,184 recombinant clones were arrayed into 326 384-well microtiter dishes. These clones were then gridded onto nylon filters (Hybond N+, Amersham Biosciences, Piscataway, USA).
[0194] The library was probed with ov...
example 3
Comparison of Genetic Structure in the Region of the Rcg1 Locus Between Resistant and Susceptible Lines and Expression Profiles of Candidate Genes Found in that Region Between Resistant and Susceptible Lines
[0201] Having found a candidate gene in the region genetically defined to carry the locus responsible for the resistance to anthracnose phenotype, efforts were undertaken first to determine if there might be other genes present in the region and second to determine if the expression patterns of the candidate gene were consistent with its putative role. Fu and Dooner ((2002), Proc Natl Acad Sci 99:9573-9578) and Brunner et al. ((2005), Plant Cell 17:343-360) have demonstrated that different corn inbred lines may have significant rearrangements and lack of colinearity with respect to each other. Comparison of such genomes over larger regions can thus be complex. Such a comparison of the genomes of Mo17 (Missouri 17) and DE811ASR (BC5) revealed that in the region where the candidat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com