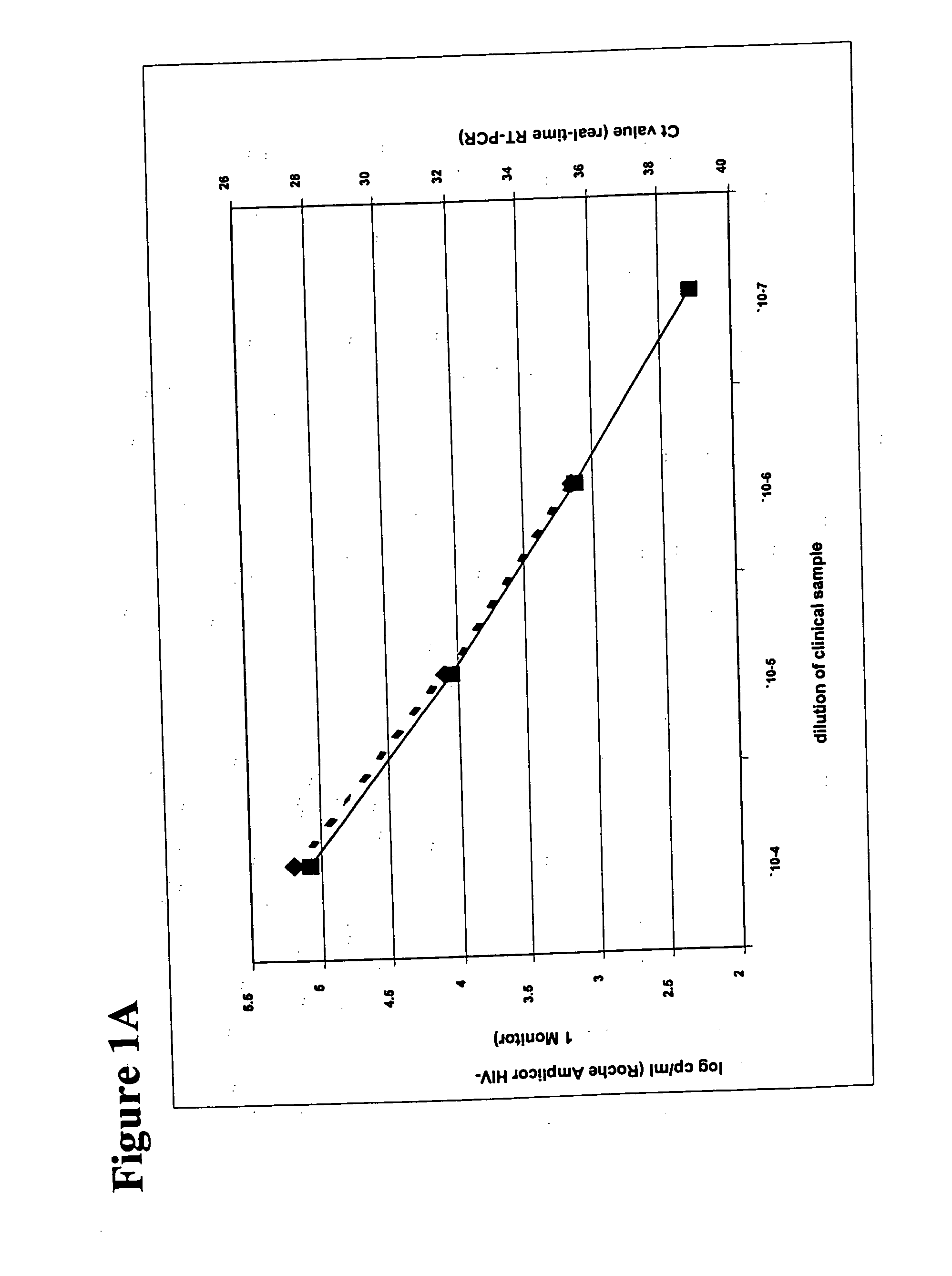
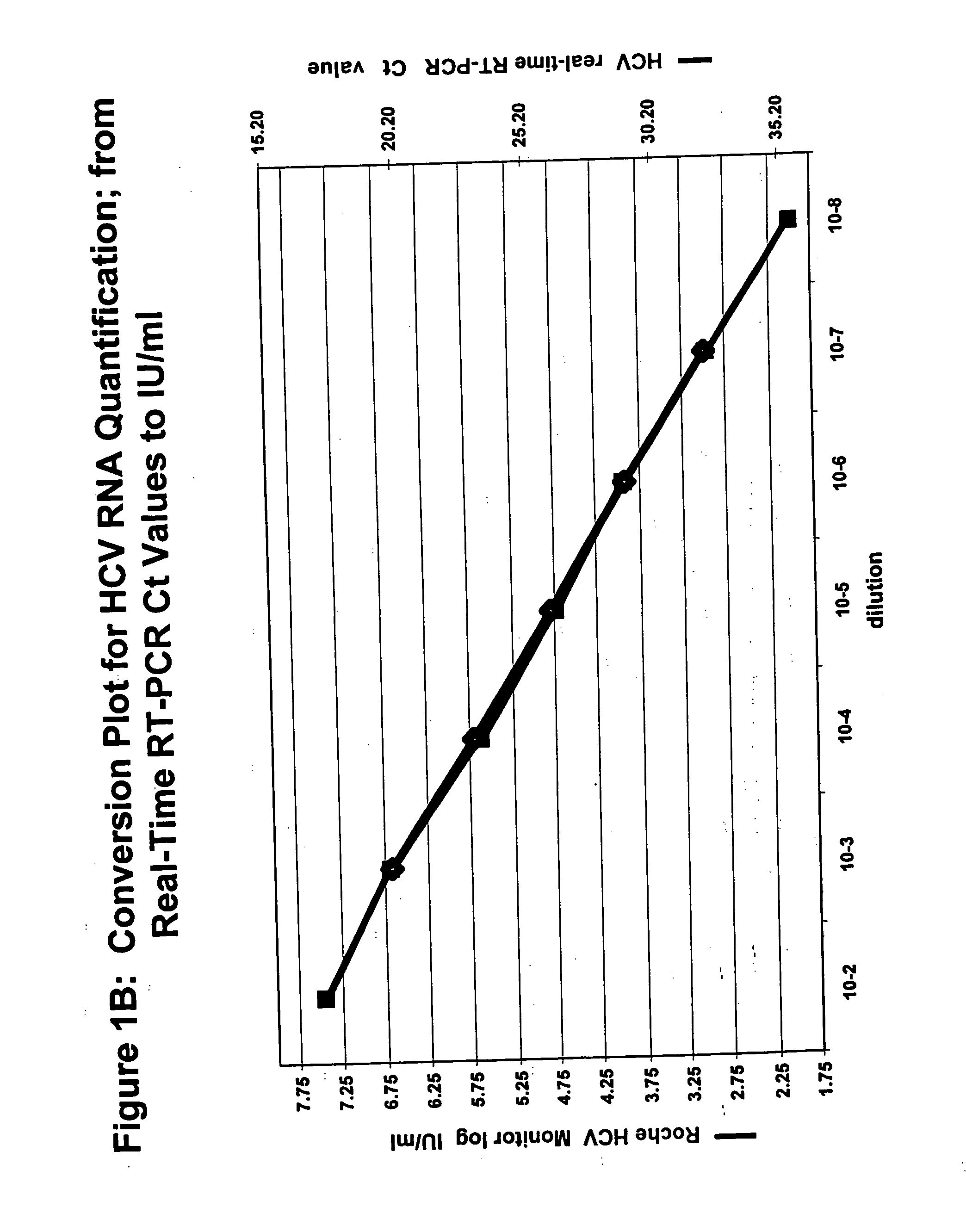
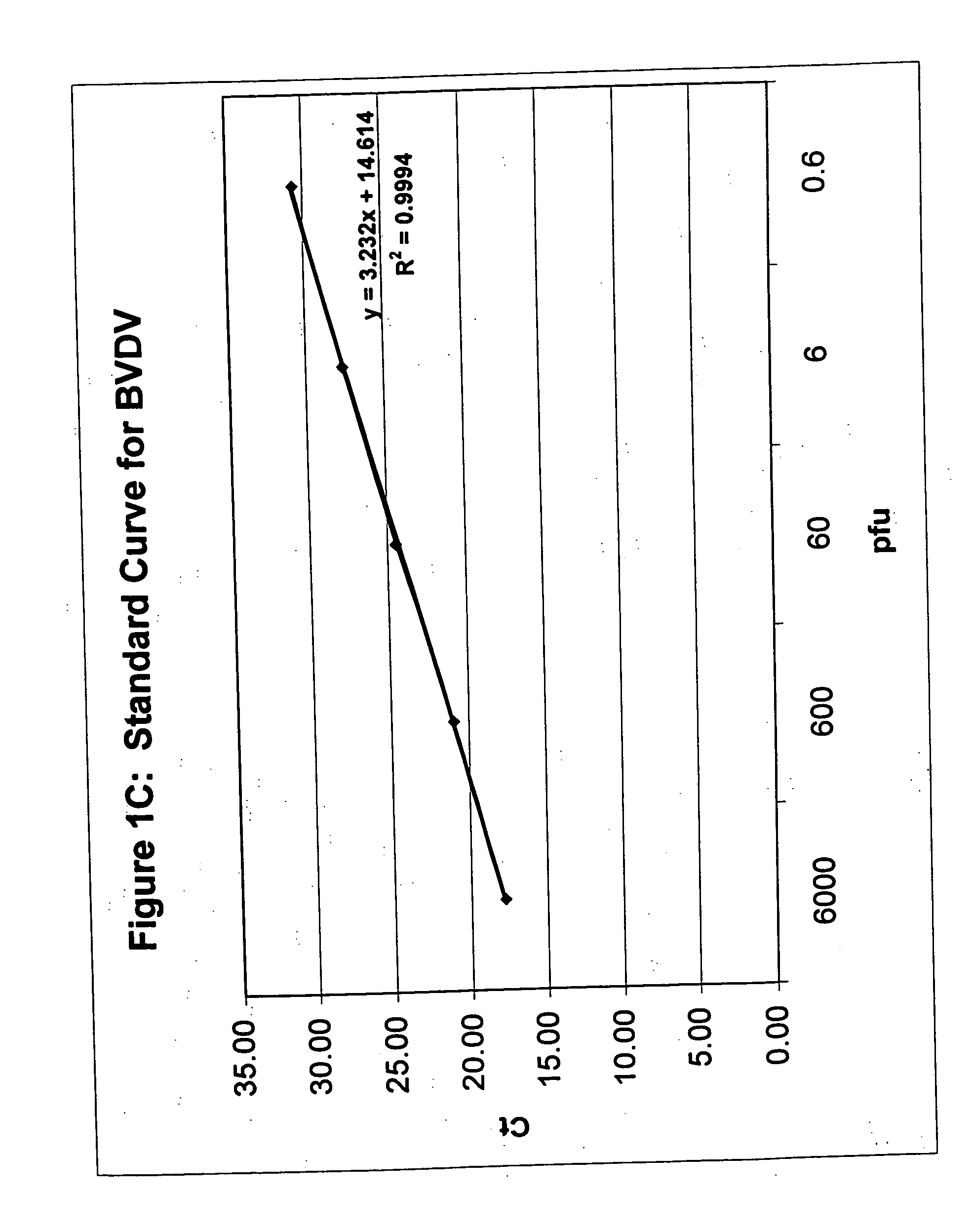
Simultaneous quantification of nucleic acids in diseased cells
a nucleic acid and nucleic acid technology, applied in the field of viral infection detection and mitochondrial toxicity, can solve the problems of more difficult sample preparation in most clinical specimens, laborious sample preparation, and trivial sample preparation, so as to assess potential side effects and eliminate variability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
HIV-1 Cell Culture
[0247] Human PBMC (1×106 cells / T25 flask) were PHA stimulated for 2 days, and infected with either a sensitive (xxBRU) or a 3TC-resistant (184V) HIV-1 strain at 100 TCID50. The culture was kept for 5 days in presence of test antiviral compounds at serial 1-log dilutions. Subsequently, human PBMC were removed from the culture supernatant by centrifugation (10 min, 400×g, 4° C.). This clarified supernatant was tested either in the RT-assay, or in the real-time RT-PCR assay.
example 2
Reverse Transcriptase (RT) Assay
[0248] Virus particles present in a 1 mL aliquot of culture supernatant were concentrated by centrifugation (2 hr, 20,000×g, 4° C.). After the 2 hour spin, supernatant fluid was removed completely and the virus pellet was dispensed into a 100 μL Virus Solubilization Buffer (VSB: 0.5% Triton X-100; 0.8 M NaCl, 0.5 mM phenylmethylsulfonyl, 20% glycerol, 50 mM Tris.HCl pH 7.8). A 10 μL aliquot of RT-VSB was mixed with 75 μL RT cocktail (60 mM Tris.HCl pH 7.8, 12 mM MgCl2, 6 mM DTT, 6 μg / mL Poly (rA)-Poly (dT), 1.2 mM dATP, and 80 μCi / mL H3-TTP) and incubated for 2 hr at 37° C. Subsequently 100 μL of 10% TCA was added, and the total amount of incorporated H3-TTP was counted.
example 3
RT-PCR Primer and Probe Assessment
[0249] The TaqMan probe and primers were designed by using the Primer Express software (Applied Biosystems, CA) and are covering highly conserved sequences complementary to the DNA sequences present in HIV-1 RNA. By scanning the different genotypes of group M for regions containing only minor variability, the conserved domain was discovered. As a result, the region in the HIV-1 RT domain between codon 200 and 280 fulfilled the required criteria; thus this region was used to design an appropriate set of primers and probe that could work in real time PCR (“RT-PCR”). Primer sequences are as follows: sense 5′-TGGGTTATGAACTCCATCCTGAT-3′ (Sequence ID No.) and 5′-TGTCATTGACAGTCCAGCTGTCT-3′ (Sequence ID No.); the probe sequence is 5′-fluorescent dye-TTTCTGGCAGCACTATAGGCTGTACTGTCCATT-quenching dye-3′ (Sequence ID No.). In this particular case, the probe was labeled with FAM at the 5′ end, and the quencher molecule is TAMARA, provided at the 3′ end. Any oth...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com