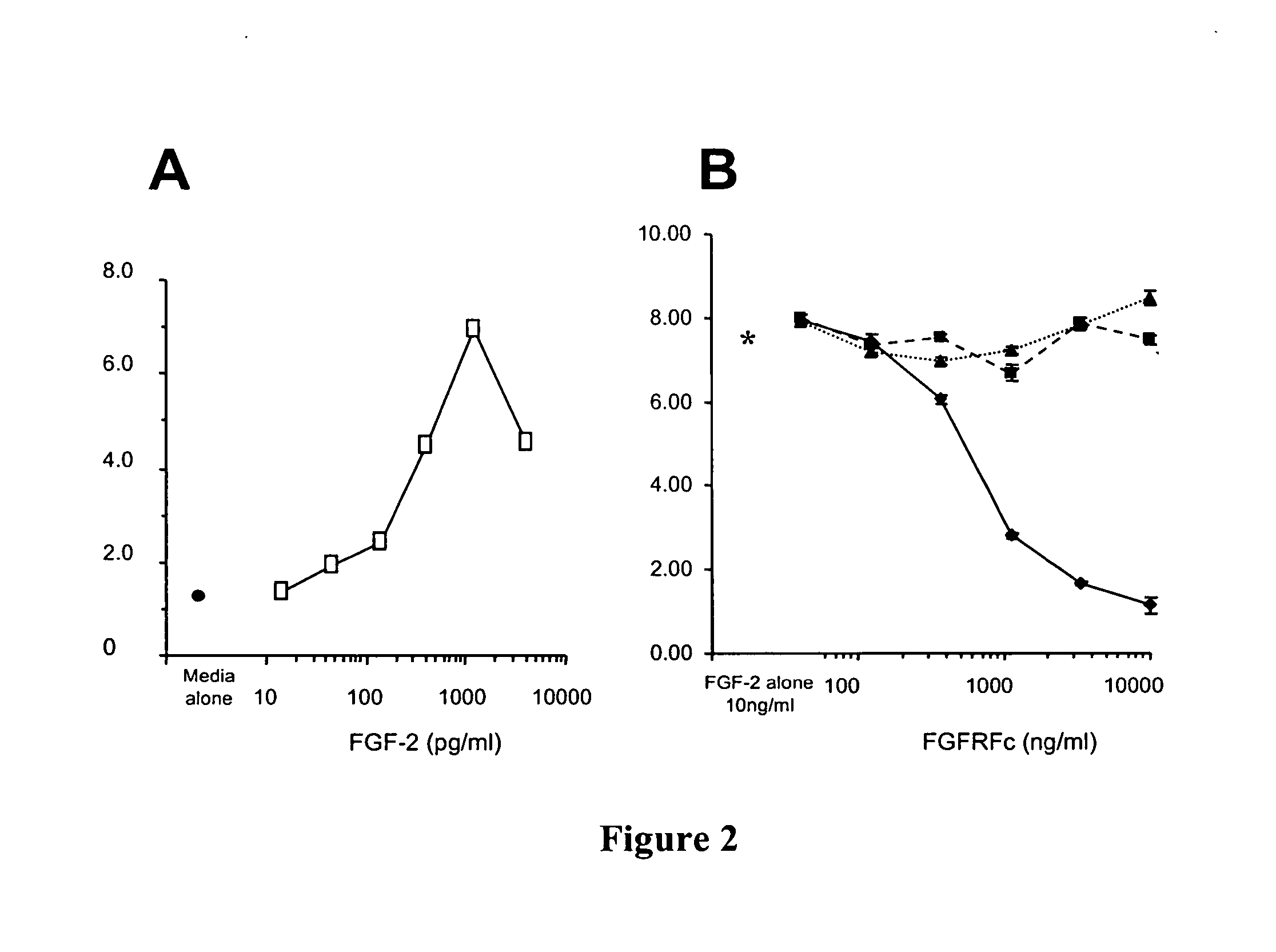
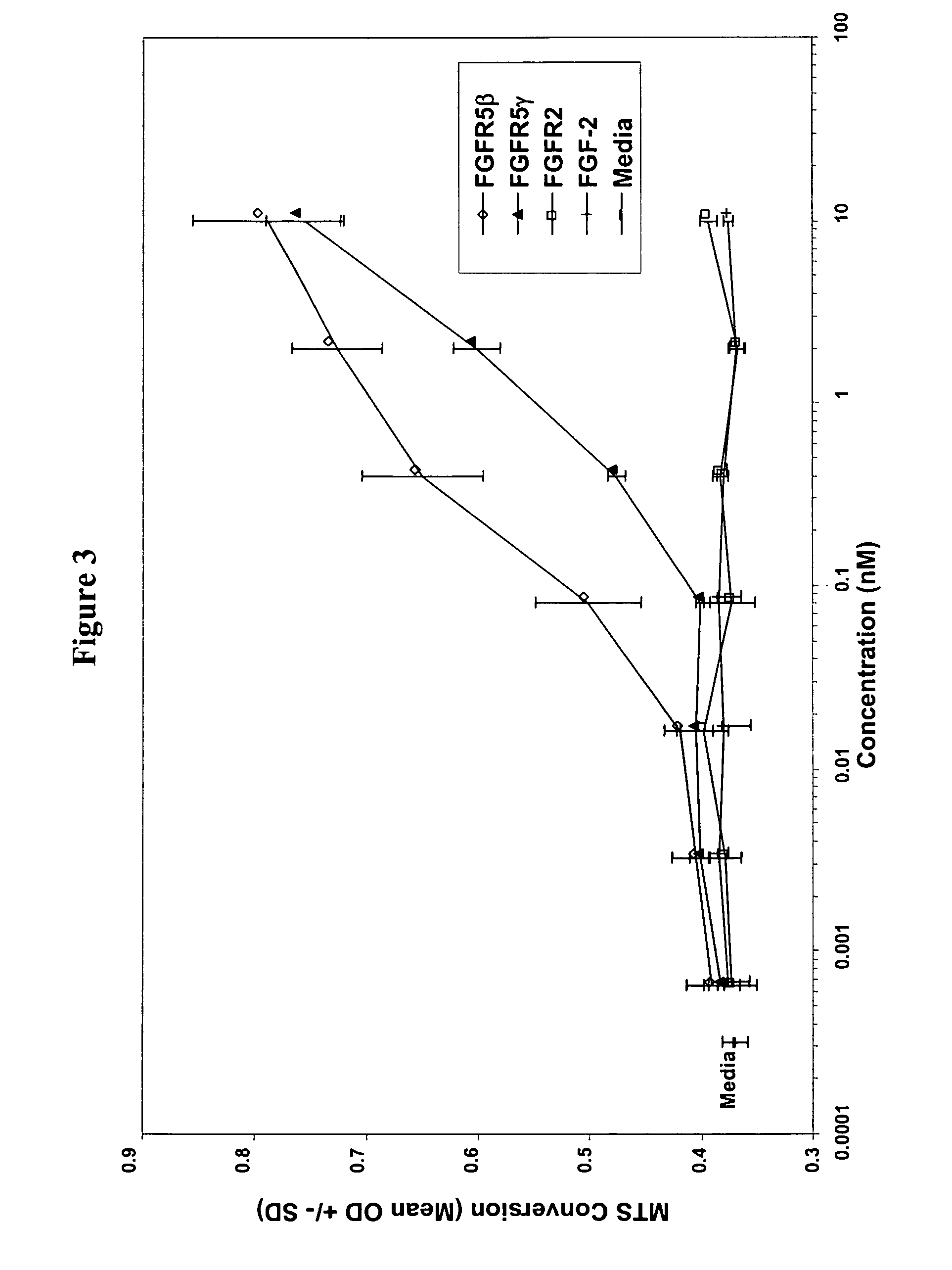
Fibroblast growth factor receptors and methods for their use
a growth factor receptor and fibroblast technology, applied in the field of fibroblast growth factor receptor 5 (fgfr5) polypeptides, to achieve the effect of decreasing fgfr5 gene expression and decreasing osteopontin gene expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of cDNA Sequences from Murine Lymph Node Stromal Cell Expression Libraries
[0156] The cDNA sequences of the present invention were obtained by high-throughput sequencing of cDNA expression libraries constructed from murine fsn- / - lymph node stromal cells as described below.
[0157] Lymph nodes were removed from flaky skin fsn- / - mice, the cells dissociated and the resulting single cell suspension placed in culture. After four passages, the cells were harvested. Total RNA, isolated using TRIzol Reagent (BRL Life Technologies, Gaithersburg, Md.), was used to obtain mRNA using a Poly(A) Quik mRNA isolation kit (Stratagene, La Jolla, Calif.), according to the manufacturer's specifications. A cDNA expression library (referred to as the MLSA library) was then prepared from the mRNA by Reverse Transcriptase synthesis using a Lambda ZAP Express cDNA library synthesis kit (Stratagene, La Jolla, Calif.). A second cDNA expression library, referred to as the MLSE library, was prepared ...
example 2
Characaterization of Isolated cDNA Sequences
[0159] The isolated cDNA sequences were compared to sequences in the EMBL DNA database using the computer algorithm BLASTN, and the corresponding polypeptide sequences (DNA translated to protein in each of 6 reading frames) were compared to sequences in the SwissProt database using the computer algorithm BLASTP. Specifically, comparisons of DNA sequences provided in SEQ ID NO: 1, 2-4 and 164 (isolated as described below) to sequences in the EMBL (Release 60, September 1999) DNA database, and the amino acid sequences correspoding to SEQ ID NO: 1-4 and 164 (provided in SEQ ID NO: 5-8, respectively) to sequences in the SwissProt and TrEMBL (up to October 20, 1999) databases were made as of Dec. 31, 1999. The cDNA sequences of SEQ ID NO: 1-4 and 164, and their corresponding polypeptide sequences (SEQ ID NO: 5-8, respectively) were determined to have less than 75% identity (determined as described above) to sequences in the EMBL and SwissProt ...
example 3
Isolation of Full Length cDNA Sequence of a Murine Fibroblast Growth Factor Receptor Homolog
[0162] The full-length cDNA sequence of a murine fibroblast growth factor receptor homolog was isolated as follows.
[0163] The MLSA cell cDNA library (described in Example 1) was screened with an [α32P]-dCTP labeled cDNA probe corresponding to nucleotides 1 to 451 of the coding region within SEQ ID NO: 1. Plaque lifts, hybridization and screening were performed using standard molecular biology techniques. The determined polynucleotide sequence of the full-length murine FGFR gene (referred to as muFGFR5β) is provided in SEQ ID NO: 2, with the corresponding polypeptide sequence being provided in SEQ ID NO: 6.
[0164] Analysis of the polynucleotide sequence of SEQ ID NO: 2 revealed the presence of a putative transmembrane domain encoded by nucleotides 1311 to 1370. The polypeptide sequence (SEQ ID NO: 6; FIG. 1) has regions similar to the extracellular domain of the fibroblast growth factor rece...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| radioactive uptake | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com