DNA-vaccines based on constructs derived from the genomes of human and animal pathogens
a technology of human and animal genomes, applied in the field of dnavaccines based on constructs derived from the genomes of human and animal pathogens, can solve the problems of non-selective expression of pathogen's genes, and achieve the effects of minimal number of molecular modifications of vectors and/or inserted polynucleotides, easy growth and manipulation, and enhanced transcription
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Nucleic Acid Immunization Using HSV-2 Cosmids
[0112] In order to assess the specificity and effectiveness of nucleic acid immunization using DNA vaccine cosmids containing HSV-2 genomic DNA, the following studies were carried out.
A. Cosmid Preparation
[0113] HSV-2 Cosmids:
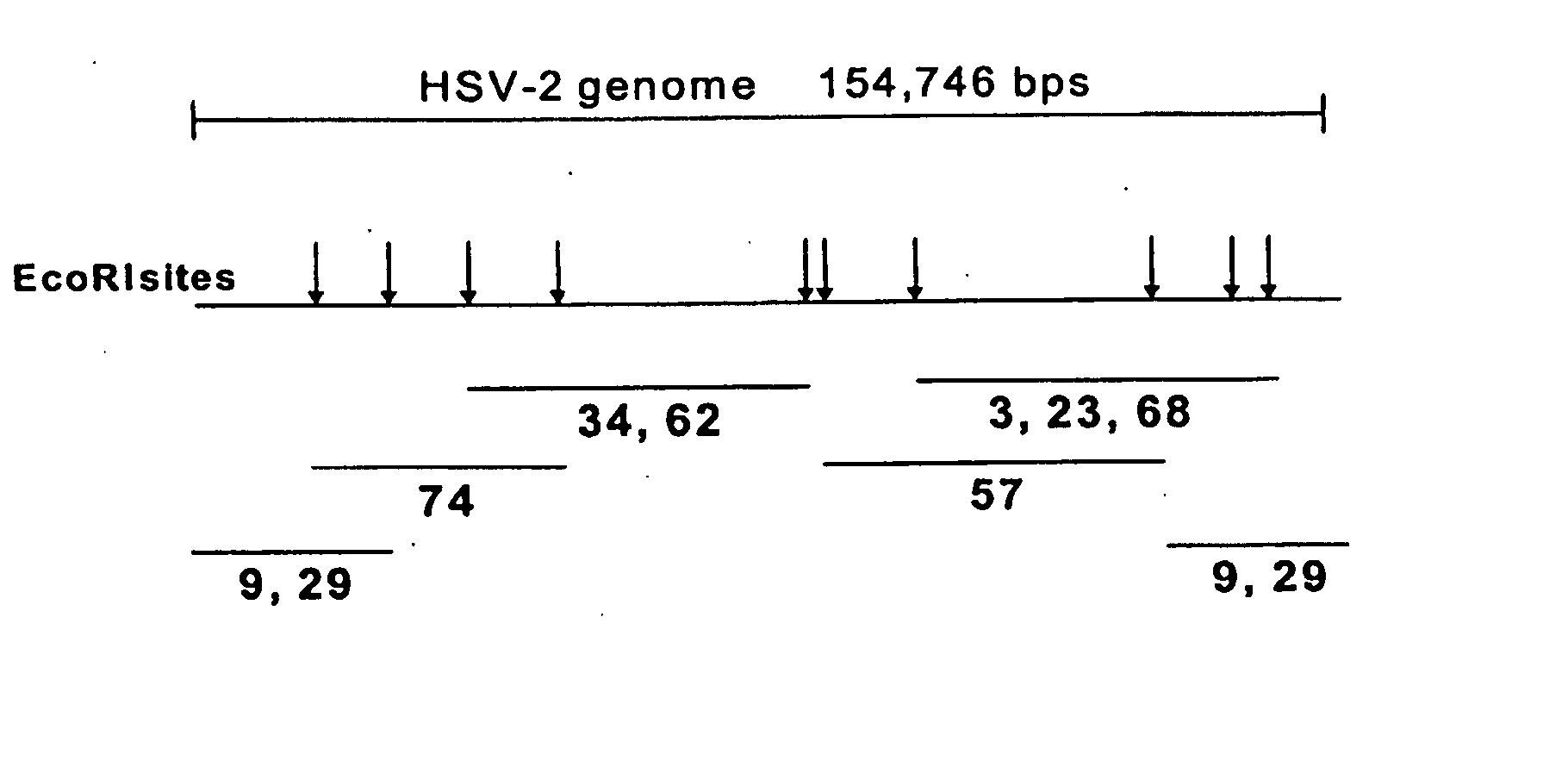
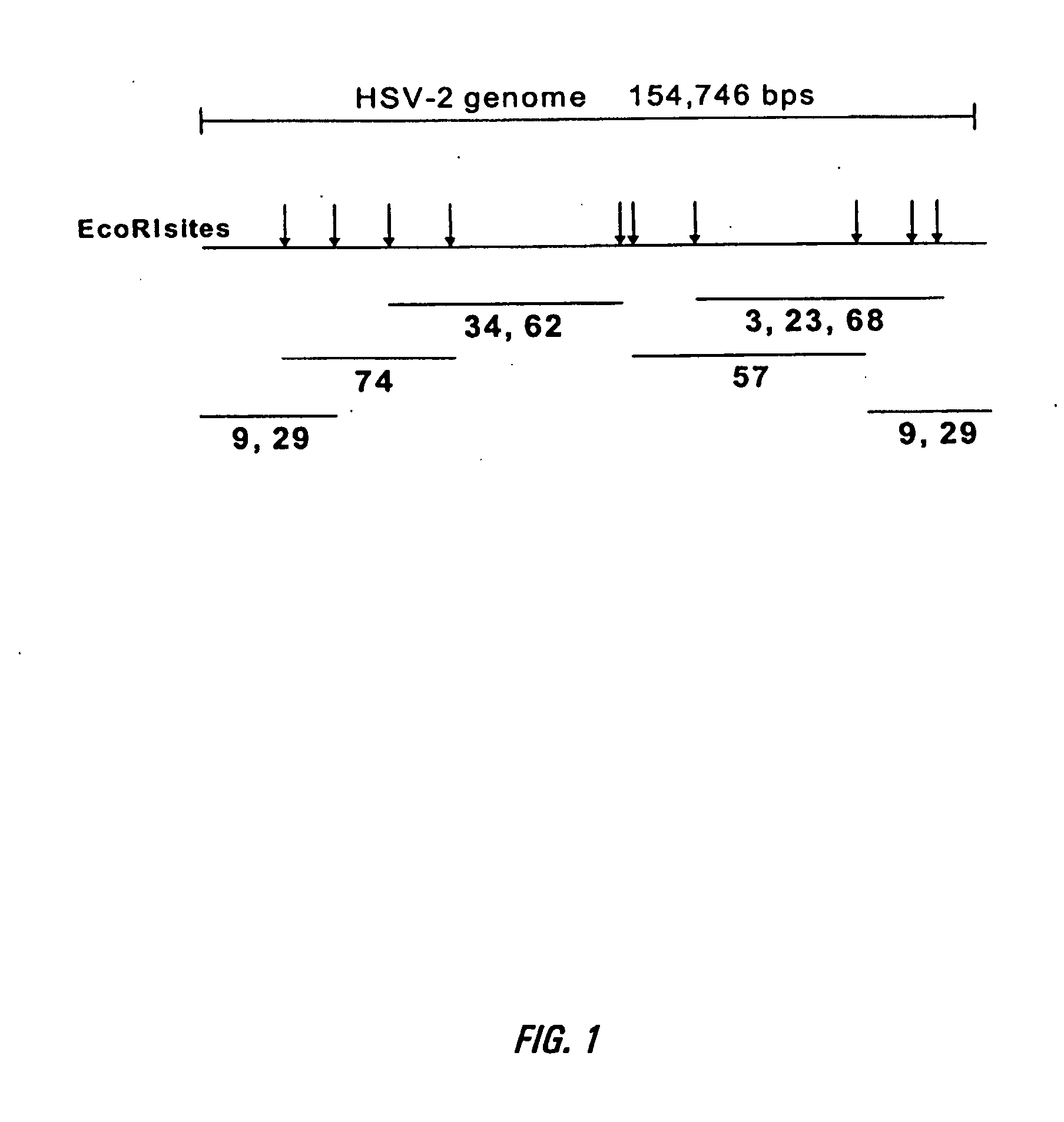
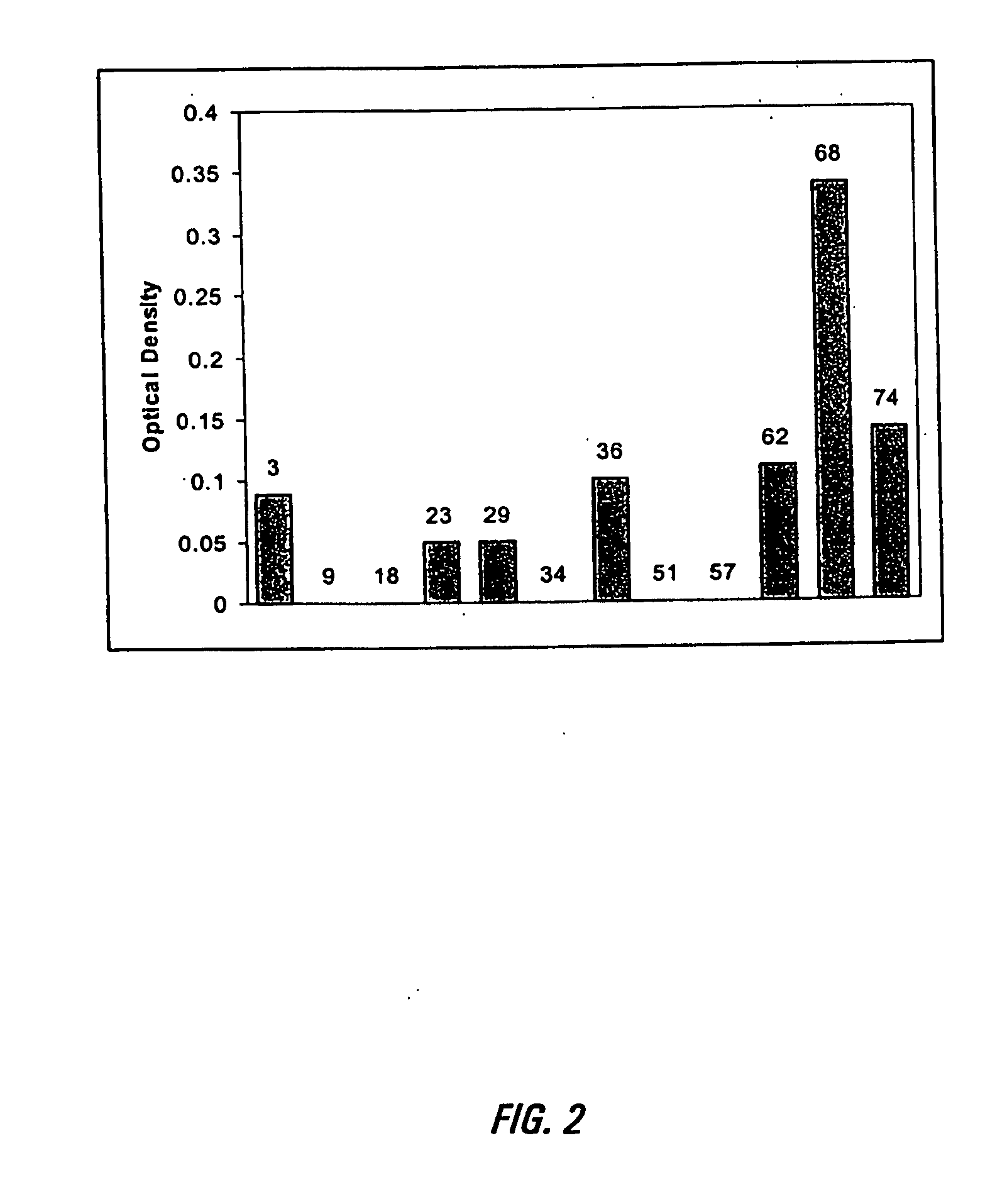
[0114] HSV-2 genomic DNA was obtained by infecting Vero cells (ATCC # CCL-81) with HSV-2 strain MS (ATCC # VR-54T) and isolating genomic DNA 10′ from the infected cells. Cosmids containing fragments of the HSV-2 genome were generated by digesting HSV-2 genomic DNA, as described above, with the restriction enzyme EcoRI (New England Biolabs). As shown in FIG. 1, the HSV genome has been mapped and contains at least 10 EcoR1 sites, indicated in the figure with arrows. The digested DNA was ligated into cosmids using the SuperCos 1 Cosmid Vector Kit from Stratagene following the manufacturers instructions. Positive cosmids were first identified from the fragment sizes generated by an EcoRI digest of purified cosmid DN...
example 2
Nucleic Acid Immunization Using HSV-2 Plasmids
A. Plasmid Construction
[0124] HSV-2 genomic DNA was obtained by infecting VERO cells (ATCC #CCL-81) with HSV-2 strain MS (ATCC #VR-54T) and isolating genomic DNA from purified viral particles. PCR was performed using the genomic DNA to amplify segments for cloning into a plasmid vector.
[0125] The primer gB2, having the following sequence: CGC GTC TAG AAA CGT TCG CGA CCA CGG GTG AC (SEQ ID NO: 3) and primer gB3, having the following sequence: CGC GTC TAG ATG ATG GGG TCC CGC TAA CTC GC (SEQ ID NO: 4), which correspond to sequences at 58160 and 52930 of the HSV-2 genome, respectively, were used to amplify a product of approximately 5200 bp by PCR. A second PCR product was generated with using primer gB2 (SEQ ID NO: 3) and primer gB4, having the following sequence: CGC GTC TAG ACC TTC ATG ACC GCG CTG GTC CT (SEQ ID NO: 5) which correspond to sequences at 58160 and 49670, respectively, of the HSV-2 genome. This produced a product of appro...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com