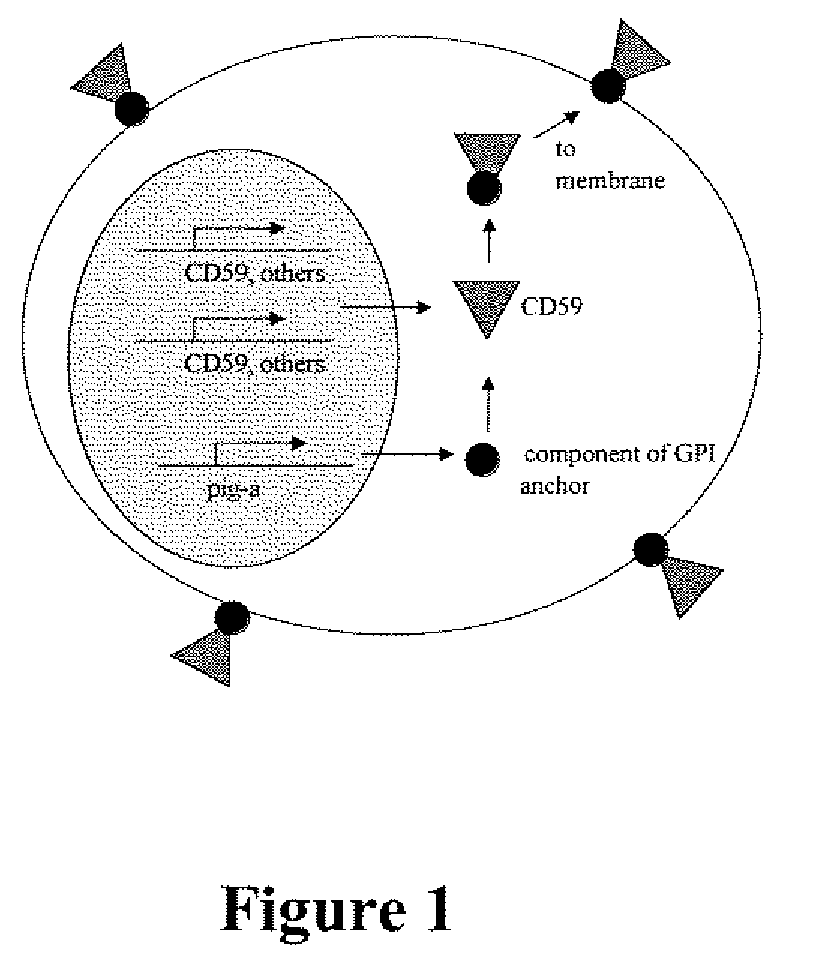
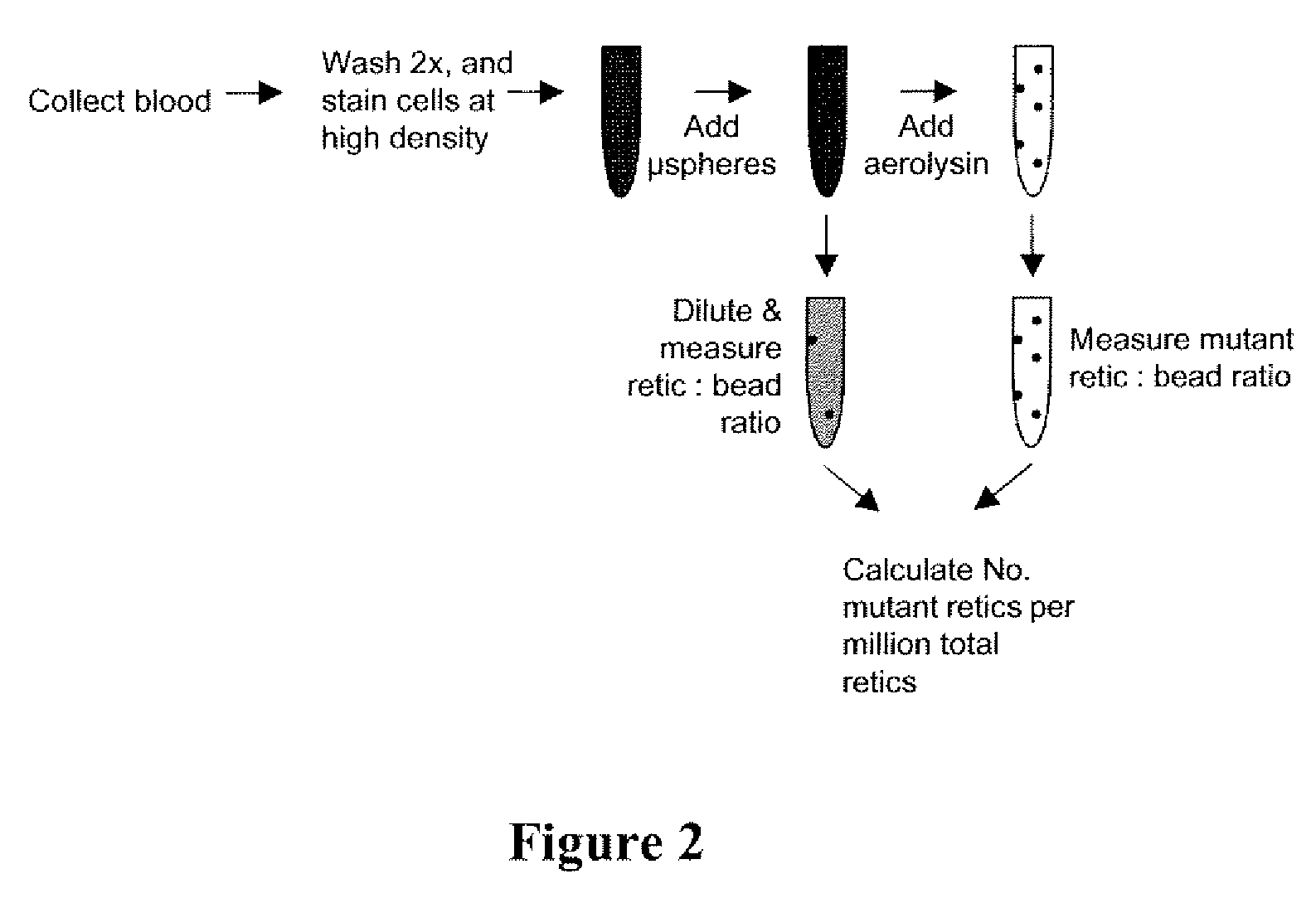
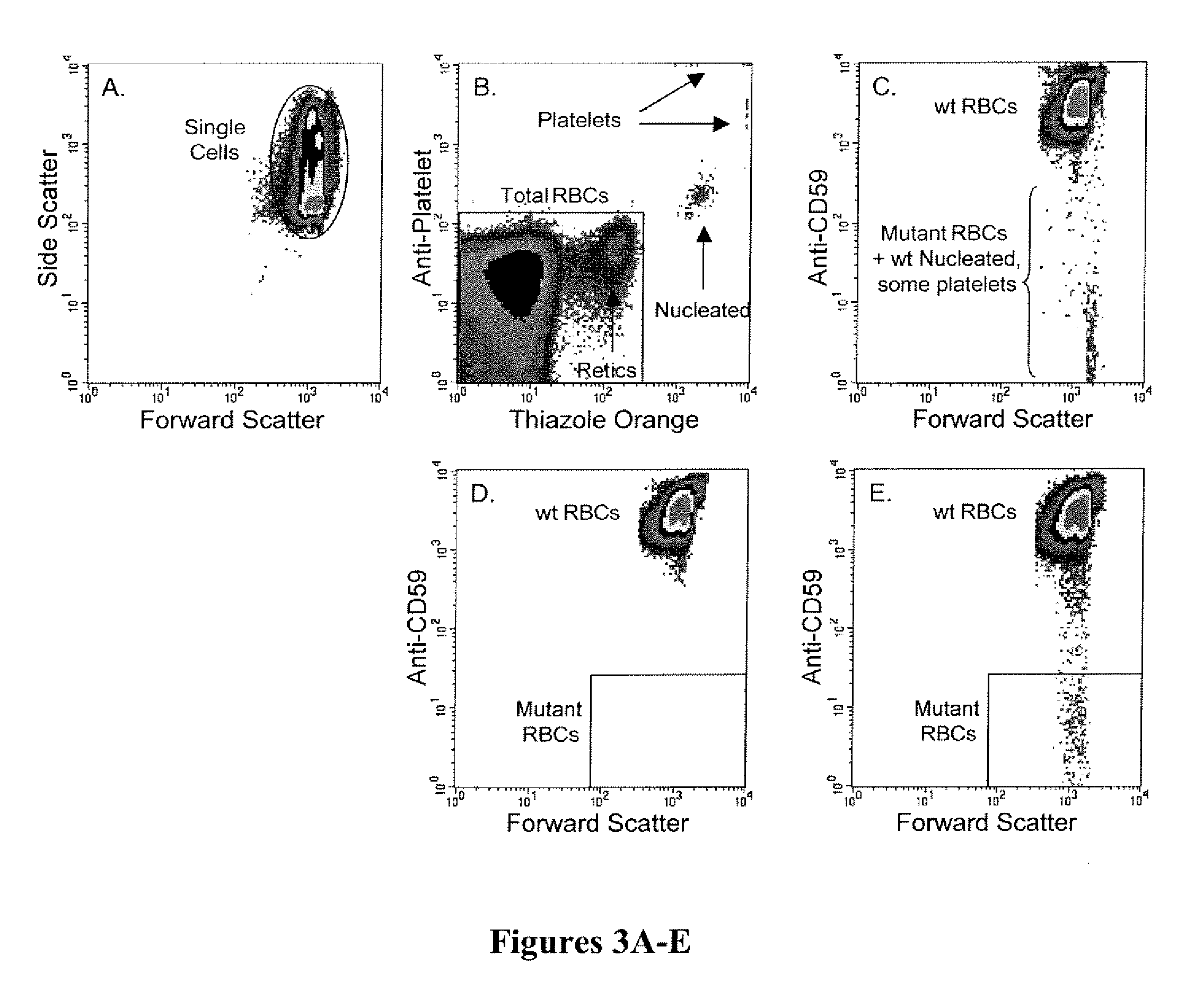
Method for measuring in vivo mutation frequency at an endogenous gene locus
a technology of endogenous gene and which is applied in the field of in vivo mutation frequency measurement at an endogenous gene locus, can solve the problems of expensive breeding programs, absence or reduction of membrane expression of gpi-linked proteins in peripheral blood cells, etc., and achieves the effect of fast, reliable and accura
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Differential Labeling of Erythrocytes, Reticulocytes, Nucleated Cells, Platelets, and Mutant Erythrocytes
[0062]In this example, a three-color procedure for differentially labeling and scoring pig-a cells is demonstrated.
[0063]Whole blood specimens (EDTA as anticoagulant) were obtained from the tail vein of vehicle- or ethyl-N-nitrosourea (ENU) treated rats (100 mg / kg / day for three days at 48 hr intervals; blood collected several weeks after the final administration). These specimens were centrifuged at low speed at least two times to significantly reduce the number of platelets. Thereafter, cells were contacted with anti-CD59-PE to differentially label mutant and non-mutant erythrocytes, and biotinylated anti-CD61 followed by streptavidin-Cy5-PE as a platelet-specific label used to exclude these particles from analysis. After washing steps to remove unbound antibodies and unbound steptavidin-Cy5-PE, cells were resupsended in a thiazole orange solution (0.1 μg / ml). Specimens were inc...
example 2
Thresholding Technique Provides Rapid Assessment of Reticulocytes for pig-a Mutant Phenotype
[0066]The thresholding technique is based on the fact that the vast majority of unfractionated blood cells are mature erythrocytes.
[0067]With appropriate staining (e.g., thiazole orange, SYTO 13 dye, SYTO 83 dye, RNASelect, or anti-CD71), it is possible to distinguish reticulocytes from their more mature counterparts.
[0068]In this case, it is possible to set the threshold parameter to the nucleic acid dye-associated fluorescence channel (e.g., FL1 for thiazole orange), as opposed to the more common forward scatter trigger. When set sufficiently high (that is, above the fluorescence intensity of mature erythrocytes), this can restrict analysis to reticulocytes.
[0069]Even so, fluorescence thresholding alone does not improve reticulocyte interrogation rates. Rather, it must be coupled with the preparation of very high density specimens. When these two adjustments are made, much quicker data acqu...
example 3
Selective Lysis of Wild-Type Erythrocytes Enhances the Speed of Mutant Phenotype Scoring
[0071]A second approach for enhancing the rate at which erythrocytes can be evaluated for the pig-a mutant phenotype is based on selective cell lysis.
[0072]To demonstrate this technique, female Sprague-Dawley rats were either untreated, or exposed to ENU at 100 mg / kg / day or 7,12-dimethyl-1,2-benz[a] anthracene (DMBA) at 40 mg / kg / day. Treatment occurred on three days (M, W, F), and tail vein blood was collected 6 weeks thereafter into EDTA-containing solutions. Blood cells were washed two times and are then stained with three fluorescent reagents according to the present invention. Specifically, specimens were contacted with anti-CD59-FITC to differentially label mutant and non-mutant erythrocytes, biotinylated anti-CD61 followed by streptavidin Cy5-PE as a platelet-specific label used to exclude these particles from analysis, and finally SYTO 83 dye to differentially stain mature erythrocytes, re...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| fluorescent emission spectrum | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com