Novgel methods for displaying (POLY)peptides/proteins on bacteriophage particles via disulfide bonds
a technology of disulfide bonds and peptides, which is applied in the direction of viruses/bacteriophages, peptide sources, fused cells, etc., can solve the problems of undeired artefacts, loss of most interesting binders which bind with high affinity to the target, and toxic expression products of gene iii to the host cell
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Display of (Poly)Peptides / Proteins on the Surface of Non-Engineered Filamentous Bacteriophage Particles via Formation of Disulfide Bonds
[0135] In the following example, all molecular biology experiments are performed according to standard protocols (Ausubel et al., 1999).
[0136] Construction of Vectors Expressing scFvs
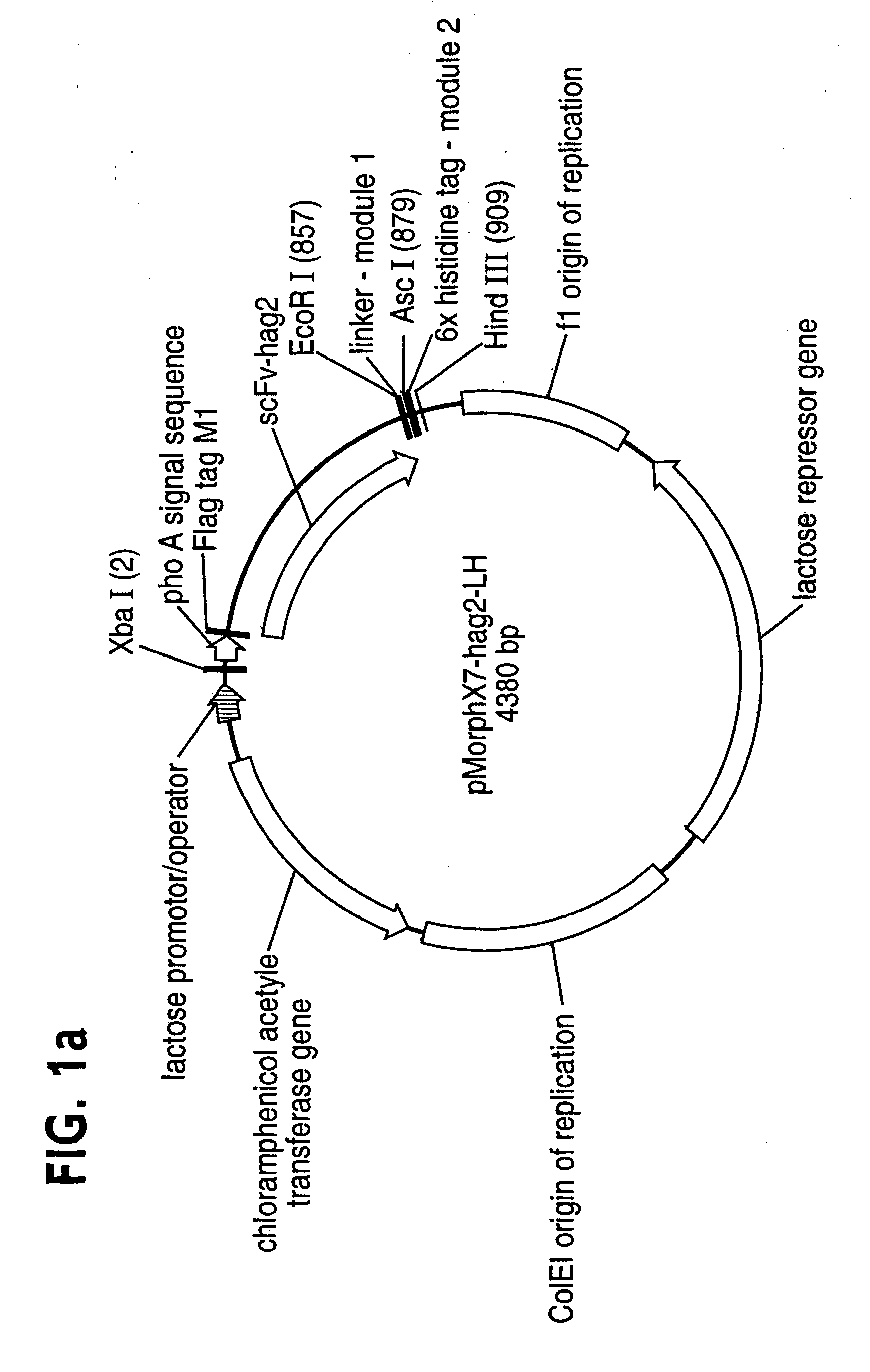
[0137] All vectors used are derivatives of the high copy phagemid pMorphX7-LH (FIG. 1a+b), a derivative of the pCAL vector series (WO 97 / 08320; Knappik et al., 2000). The expression cassette comprises the phoA signal sequence, a minimal binding site for the monoclonal antibody (mab) anti-FLAG M1 (Sigma #F-3040) (Knappik and Plückthun, 1994), a single chain fragment (scFv), a short linker (PGGSG) and a 6x histidine tag (6His; Hochuli et al., 1988) (FIG. 1a). pMorphX7-LCH and pMorphX7-LHC have been generated by inserting oligonucleotide cassettes coding for Cys-6His and 6His-Cys, respectively, between the unique AscI and HindIII sites of pMorphX7-LH (FIG. 1a, Table 1)....
example 2
Display of (Poly)Peptides / Proteins on the Surface of Engineered Filamentous Bacteriophage Particles via Formation of Disulfide Bonds
example 2.1
Display of scFvs
[0146] Example 1 described above shows that functional scFvs can be displayed on non-engineered phages via disulfide bonds. This system can be further improved, e.g. via engineering an exposed cysteine on a phage coat protein. One candidate phage coat protein is protein III (pIII) which is composed of three domains N1, N2 and pMCT. Possible sites for positioning an unpaired cysteine residue are the linker regions between the domains or the exposed N-terminus of the domain or the pIIICT in a truncated pIII version. A further example would be phage coat protein IX (pIX) where the cysteine could e.g. be linked to the N-terminus of the full length protein. In principle the cassettes for expression of such engineered proteins can be placed on the vector which is providing the scFv (one-vector system), or on a separate vector (two-vector system).
[0147] In the following we will describe experiments in which we engineered both a full length and a truncated pIII version as ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com