Induction of apoptosis and cell growth inhibition by protein 4.33
a technology of cell growth inhibition and apoptosis, which is applied in the field of induction of apoptosis and cell growth inhibition by protein 4.33, can solve the problems of inability to predict the nature, size, number, absolute specificity or complexity of such postulated cell surface association proteins, and cannot be fully understood
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
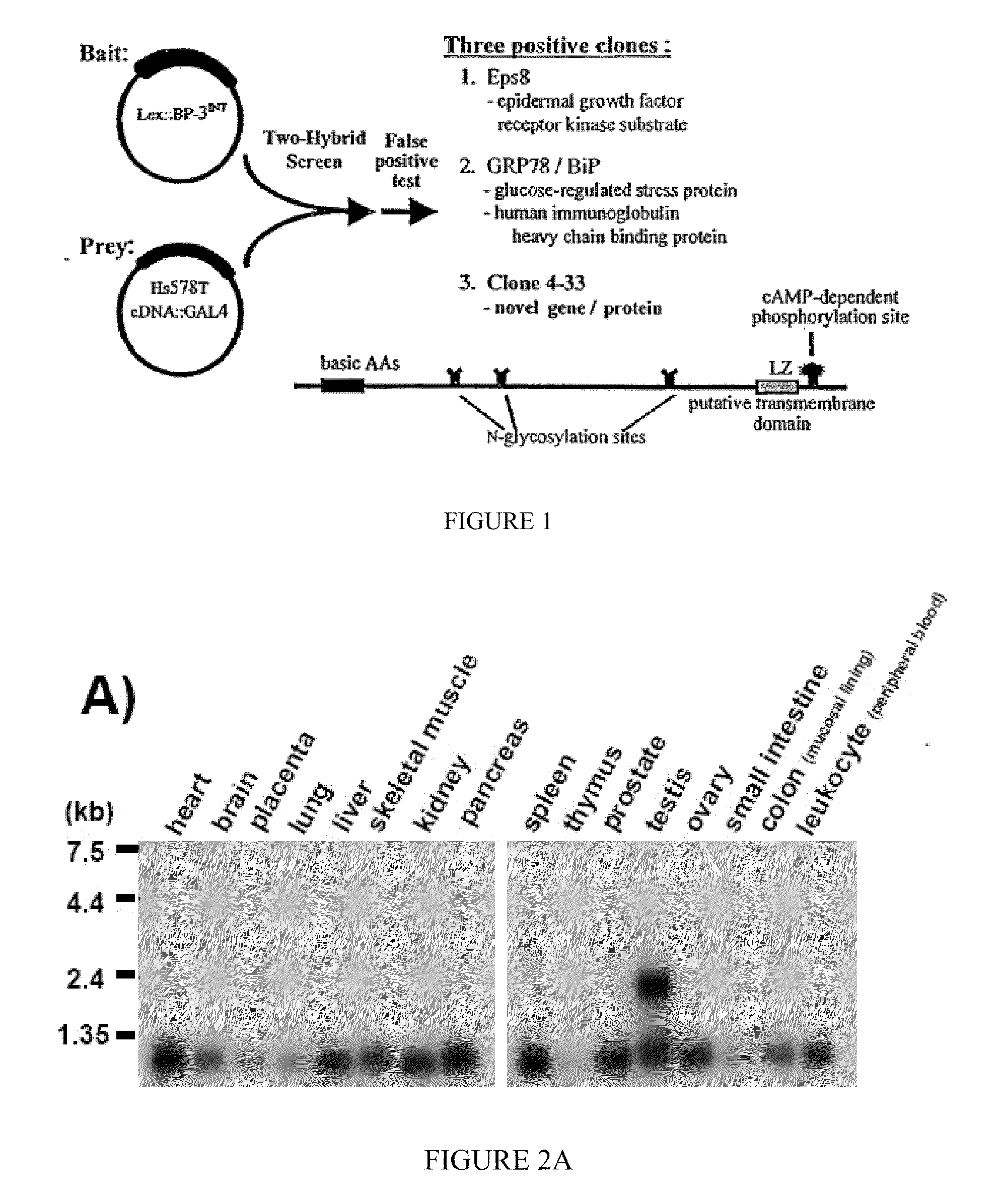
Cloning of a cDNA for a Novel IGFBP-3 Interacting Protein, P4.33
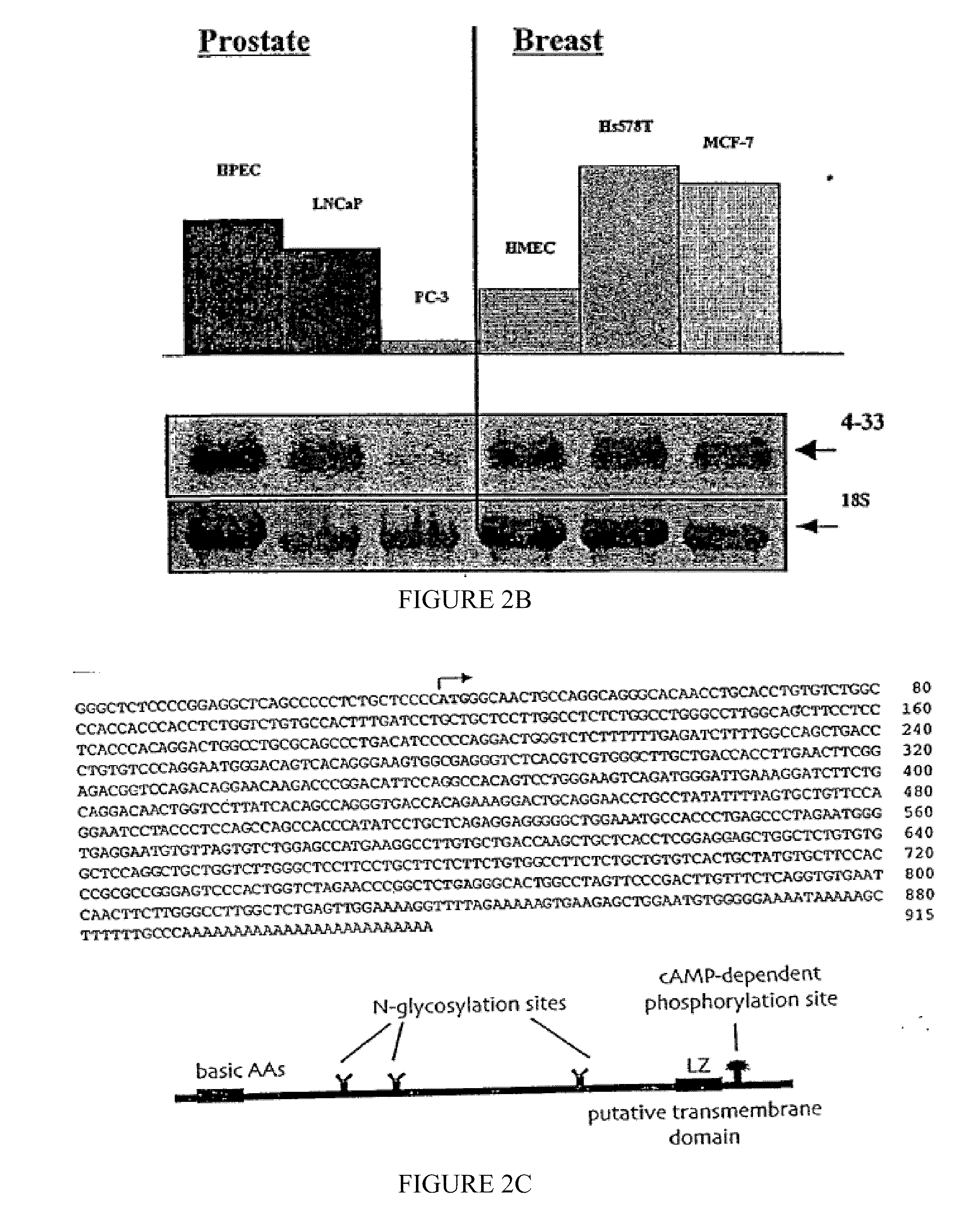
[0192]The following example of the present invention describes the cloning and isolation of clone 4.33, a cDNA encoding a novel IGFBP-3 interacting protein. The results show the discovery of a novel cDNA, 915 by in length (SEQ ID NO:1), encoding a novel 240 amino acid protein (SEQ ID NO:2) that contains several N-glycosylation sites, a cAMP-dependent phosphorylation site, a single leucine-zipper motif (LZ), and a putative transmembrane domain near the C-terminus. See FIG. 2C.
Material and Methods
[0193]Yeast two-hybrid screen. An Hs578T cDNA library in pGAD10 was generated using the Two-Hybrid cDNA Library Construction Kit (Clontech). The “bait” plasmid was constructed by amplifying an internal sequence of the IGFBP-3 cDNA (encoding amino acids 88-148) by PCR, then cloning this fragment into the pBTM116 vector, in frame with the LexA DNA binding domain coding sequence. The bait plasmid was then transformed into yeast stra...
example 2
Cellular Localization of EGFP::4.33 Fusion Protein, and Cellular Colocalization and Co-Immunoprecipitation of IGFBP-3 and Clone 4.33
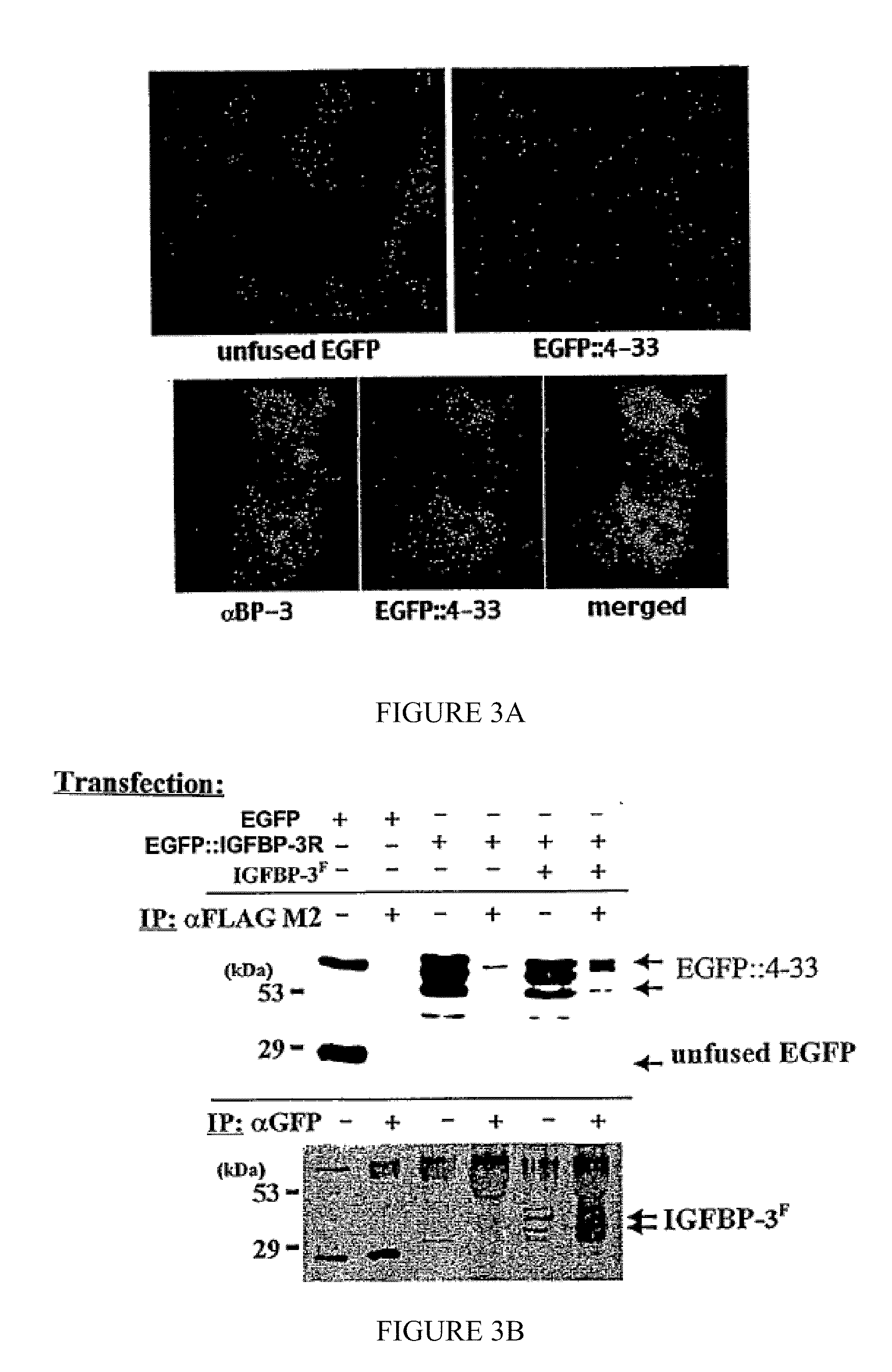
[0202]According to the present invention, the interaction between IGFBP-3 and clone 4.33 was confirmed by immunocytochemistry (FIG. 3A) and in coimmunoprecipitation studies (FIG. 3B). COS-7 cells were transiently transfected with or without constructs encoding an EGFP::4.33 fusion protein and FLAG-tagged IGFBP-3 (IGFBP-3F).
Material and Methods
[0203]Immunocytochemistry. Cells were seeded in 8-chamber culture slides (Nunc) and transfected at 70-80% confluency. After 48 hours, adherent cells were washed with PBS, fixed in 4% paraformaldehyde for 10 minutes at room temperature, and washed a further 3 times with PBS. Cells were then incubated in blocking solution (1% normal goat serum in PBS, 0.1% Triton X-100) for 1 hour at room temperature, followed by incubation with primary antibody diluted 1:1000 in blocking solution for 1-2 hours at room temperature. C...
example 3
Cellular Overexpression of Clone 4.33 Resulted in Increased Specific IGFBP-3 Cell-Surface Binding, and Co-Translocation to the Nucleus
[0212]According to the present invention, the binding of 125I-labelled IGFBP-3 to cells previously transfected with clone 4.33 was measured to determine whether the resulting expressed P4.33 could facilitate specific binding of IGFBP-3 to the cells (FIG. 4A).
Material and Methods
[0213]Cell culture, transfection, cell lysates, membrane preps. All cell lines, except the MCF-7:BP-3 stable lines, were purchased from ATCC. All were cultured in DMEM / high glucose with 10% fetal bovine serum. MCF-7:BP-3 stable lines were generated from wild type MCF-7 cells using the Ecdysone-inducible expression system (Invitrogen) according to the manufacturers instructions, and were maintained in DMEM / high glucose, 10% fetal bovine serum with 100 μg / mL Zeocin+800 μg / mL G418. IGFBP-3 protein production was induced with the addition of ponasterone A (Invitrogen) to the cultur...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com