Method and rapid test for detection of specific nucleic acid sequences
a technology of specific nucleic acid sequences and test kits, which is applied in the direction of microorganism testing/measurement, biochemistry apparatus and processes, etc., can solve the problems of insufficient use of visualization of pcr products by means of gel electrophoresis, inability of probes to function as primers during elongation, and inability to distinguish between correct products and amplification artifacts by intercalating dyes, etc., to achieve the effect of rapid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
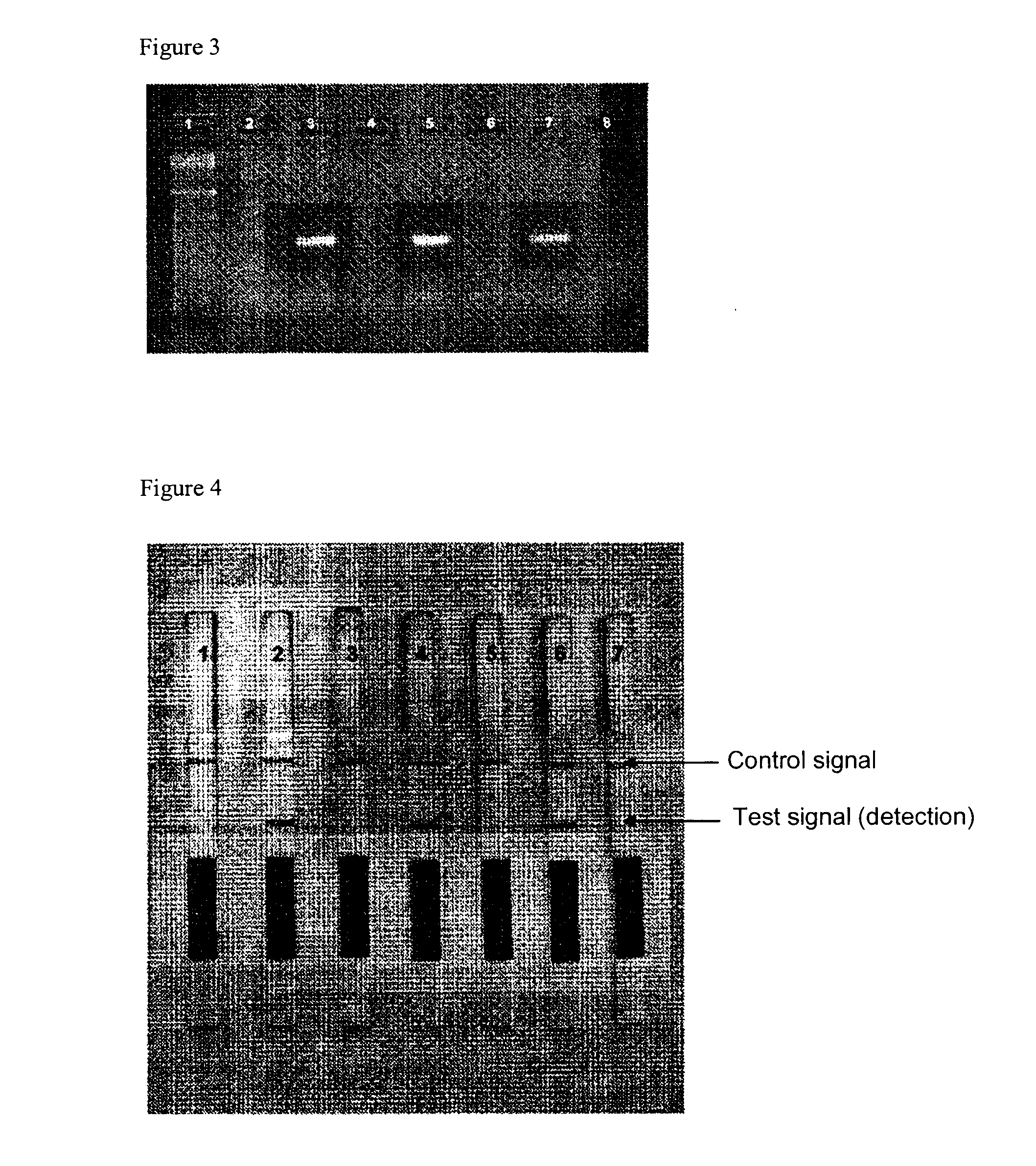
Detection of Listeria monocytogenes DNA by Means of the Hybridization Method Integrated into the PCR and of Lateral Flow Detection. Comparison of an Unphosphorylated and a Phosphorylated Probe
[0057]Two types of labeled probes were tested against one another in the mixture. The first probe is FITC-labeled at the 5′-end, and the second probe is also singly phosphorylated at its 3′-end. The 3′-phosphorylation of the probe prevents it from being elongated by the Taq polymerase.
Mixture 1 (unphosphorylated hybridization probe)
PCR primer / probe
L. monocytogenes sense primer(SEQ ID NO: 1)(5′-CGC AAC AAA CTG AAG CAA AGG-3′)L. monocytogenes antisense primer(SEQ ID NO: 2)(5′-BIOTIN-TCC GCG TGT TTC TTT TCG AT-3′)L. monocytogenes probe(SEQ ID NO: 3)(5′-FITC-CCA TGG CAC CAC CAG CAT CT-3′)
Reaction mixture (amplification / hybridization)
Per sample:
sense primer (50 pmol / μL)0.1 μLantisense primer (50 pmol / μL)0.1 μLprobe (25 pmol / μL)0.1 μLdNTP Mix (12.5 mM)0.3 μL10X PCR buffer (MgCl2 included)1.5 μLTaq-DN...
example 2
Performance of the Method by Means of Asymmetric PCR and Check of Specificity of the Test on the Basis of Testing of Positive and Negative Starting Samples
[0066]The inventive method was used as an example for detection of Rickettsia DNA isolated from tick tissue. The specificity of the method was determined by means of parallel tests on Rickettsia-negative DNA samples, also isolated from tick tissue.
PCR primer probe:
Rickettsia sense primer:(SEQ ID NO: 5)5′-GGG ACC TGC TCA CGG CGG-3′Rickettsia antisense primer:(SEQ ID NO: 6)5′-Biotin-TCT ATT GCT ATT TGT AAG AGC GGA TTG-3′Rickettsia probe:(SEQ ID NO: 7)5′-FITC- CAA AGA AGT ATT AAA GGA ACT C-Pho-3′
Reaction mixture (amplification / hybridization)
Per sample:
sense primer (50 pmol / μL)0.05 μL antisense primer (50 pmol / μL)0.1 μLprobe (25 pmol / μL)0.1 μLdNTP Mix (12.5 mM)0.3 μL10X PCR buffer (MgCl2 included)1.5 μLTaq-DNA polymerase0.75 U DNA (positive or negative)1.5 μL (approx. 50 ng)PCR-grade H2Oadd 15 μl
[0067]The PCR was performed in the Spee...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com