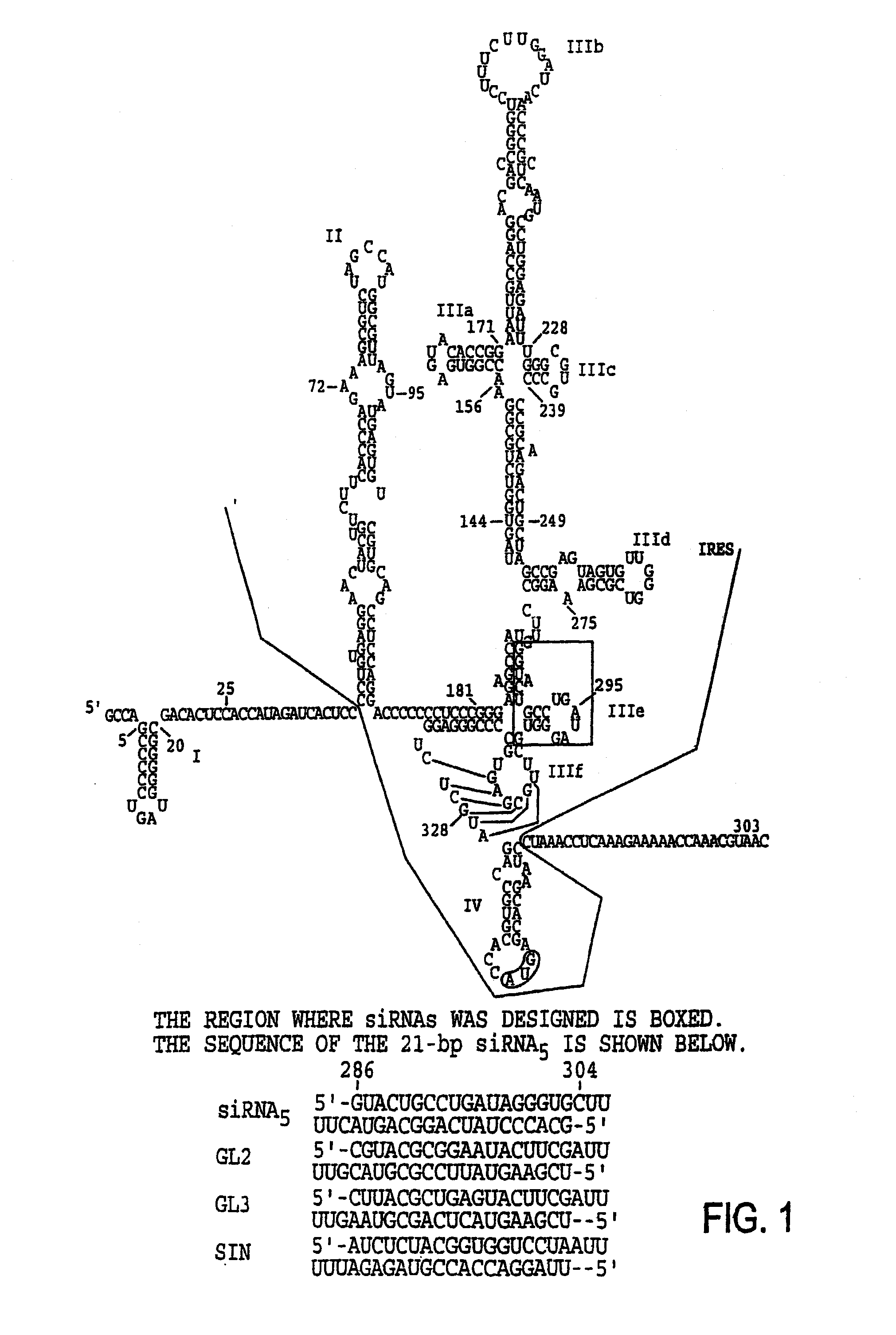
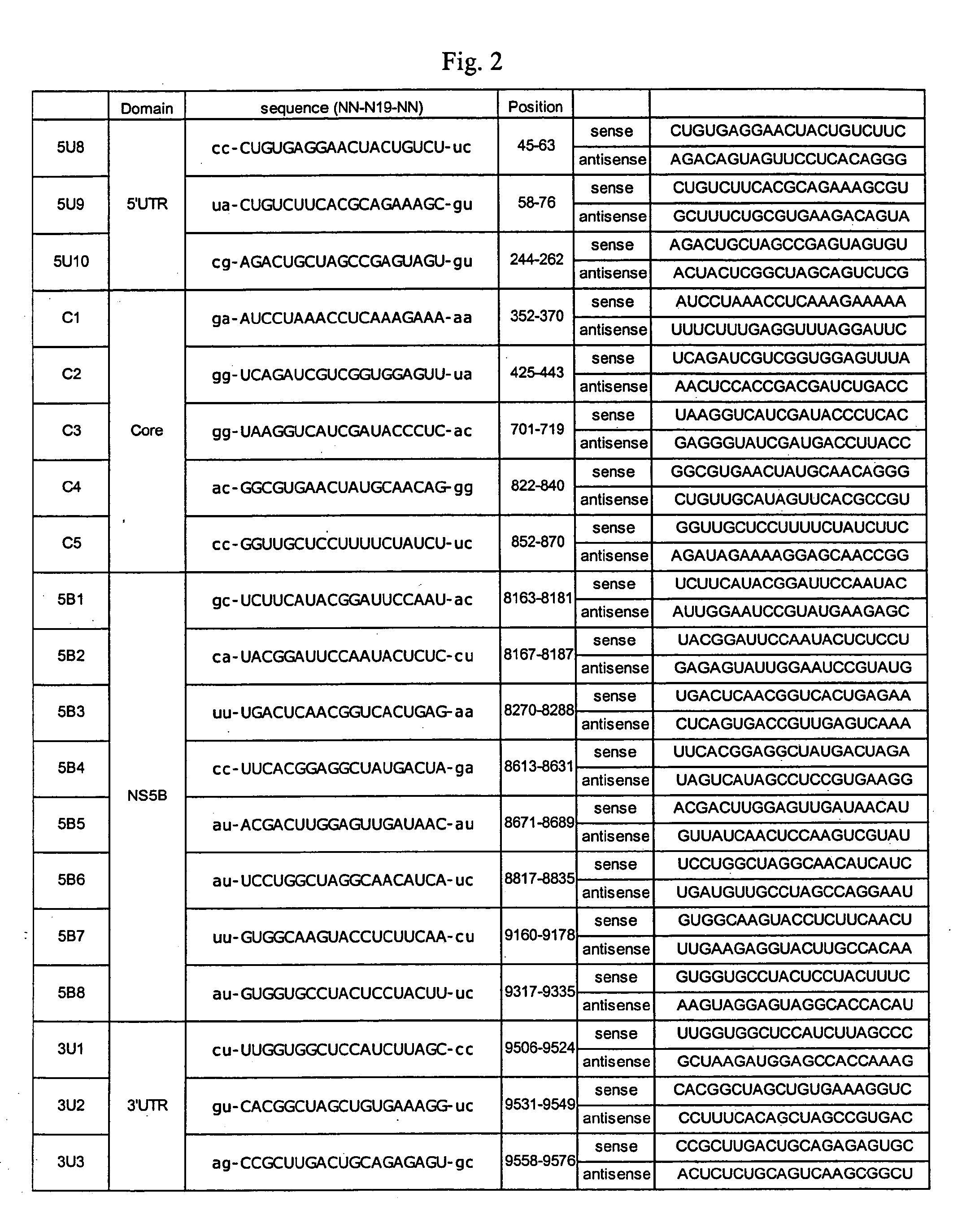
Modified Small Interfering RNA Molecules and Methods of Use
a technology of rna interference and small molecules, applied in the field of nucleic acid detection and to the phenomenon of rna interference, or rna interference, can solve the problems of inability to determine the sequence of a mammalian cell type's genome, inability to determine whether every mammalian cell type possesses the necessary machinery, and inability to achieve long-term effects. , to achieve the effect of inhibiting the replication of hcv
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0107]To test whether siRNA directed to the HCV genome confers intracellular immunity against this human pathogen, a recently developed HCV cell culture systems in human hepatoma cell line, Huh-7, was used. One of the cell lines, 5-2, harbors autonomously replicating subgenomic HCV RNA (Bartenschlager, 7. Virol, 2001). The subgenomic replicon carries firefly luciferase gene, allowing a reporter function assay as a measure of HCV RNA replication (FIG. 5). Owing to cell culture adaptive mutations introduced into the genome (Bart), these 5-2 cells replicate HCV RNA at levels of up to 5×104 virus particles / cell.
[0108]Using T7 transcription, several 21-bp siRNA duplexes against different regions of the 5′-UTR of the HCV genome were made (FIG. 5). Briefly, 2 oligo double-stranded DNA molecules comprising the T7 promoter and the 5′ UTR of HCV being oriented in either the sense direction or the antisense direction were generated. Each oligo DNA was then transcribed in vitro to produce (+) a...
example 2
[0110]The sequence specificity of the siRNA5 response was further tested using additional siRNA duplexes, GL2 and GL3. FIG. 1 shows that GL2 and GL3 differ from each other by 3-nucleotides. Luciferase activity was reduced by 90% in cells transfected with siRNA5 or GL2, but no significant reduction was seen in cells transfected with GL3 (FIG. 7). The luciferase assay was performed using a Luciferase assay system available from Promega Corp. (Madison, Wis.), according to the manufacturer's instructions.
example 3
[0111]Whether or not siRNA5 was toxic to transfected cells also was tested. Toxicity was by measured using an ATPase activity assay. FIG. 8 shows that the siRNA5-induced reduction in HCV replication, as seen in FIG. 6, was not due to cellular toxicity which is attributed to non sequence-specific RNAi. ATPase levels were assayed using an ATPase assay kit from Promega (Madison, Wis.) according to the manufacturer's instructions.
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com