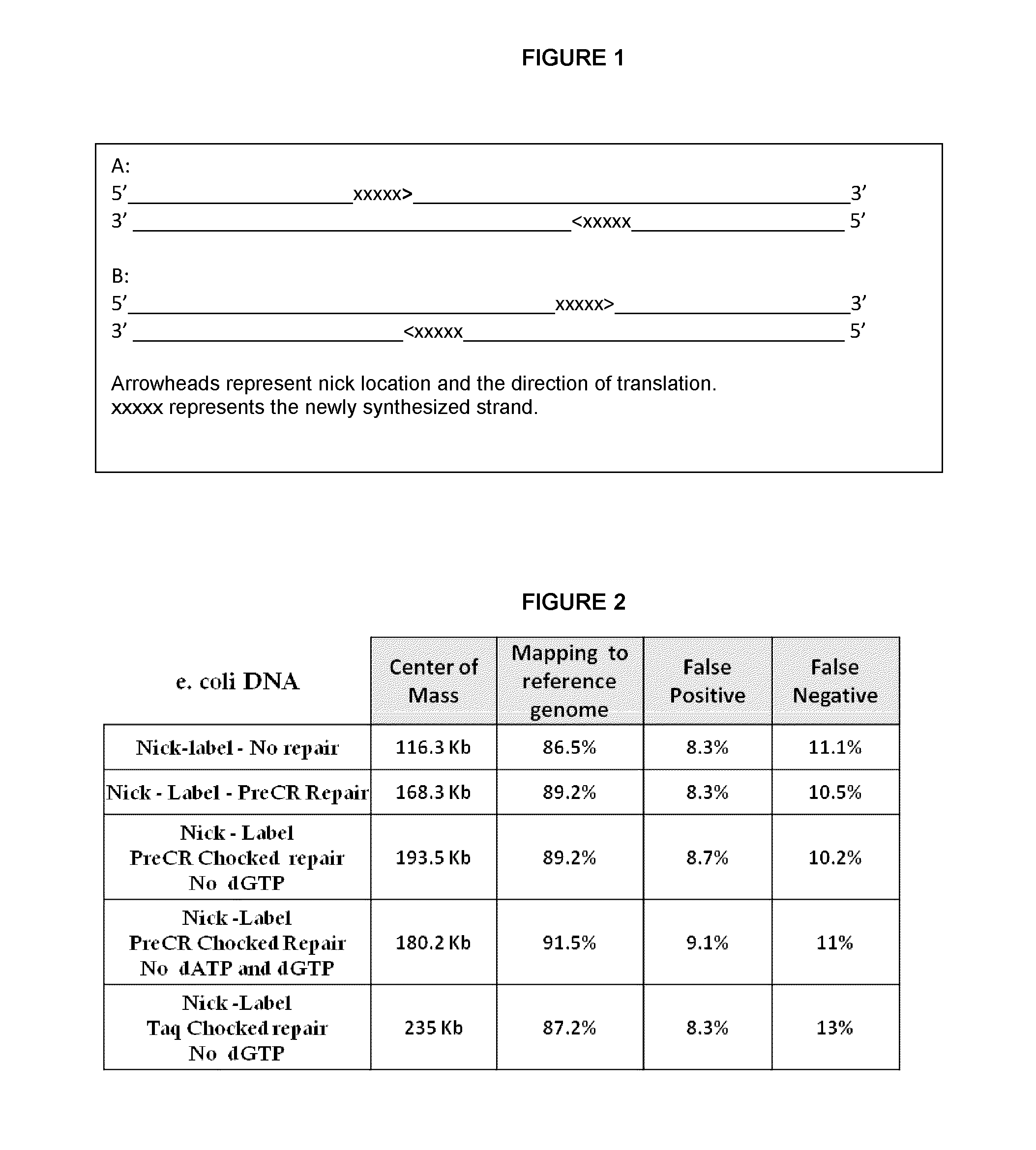
Methods for single-molecule analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
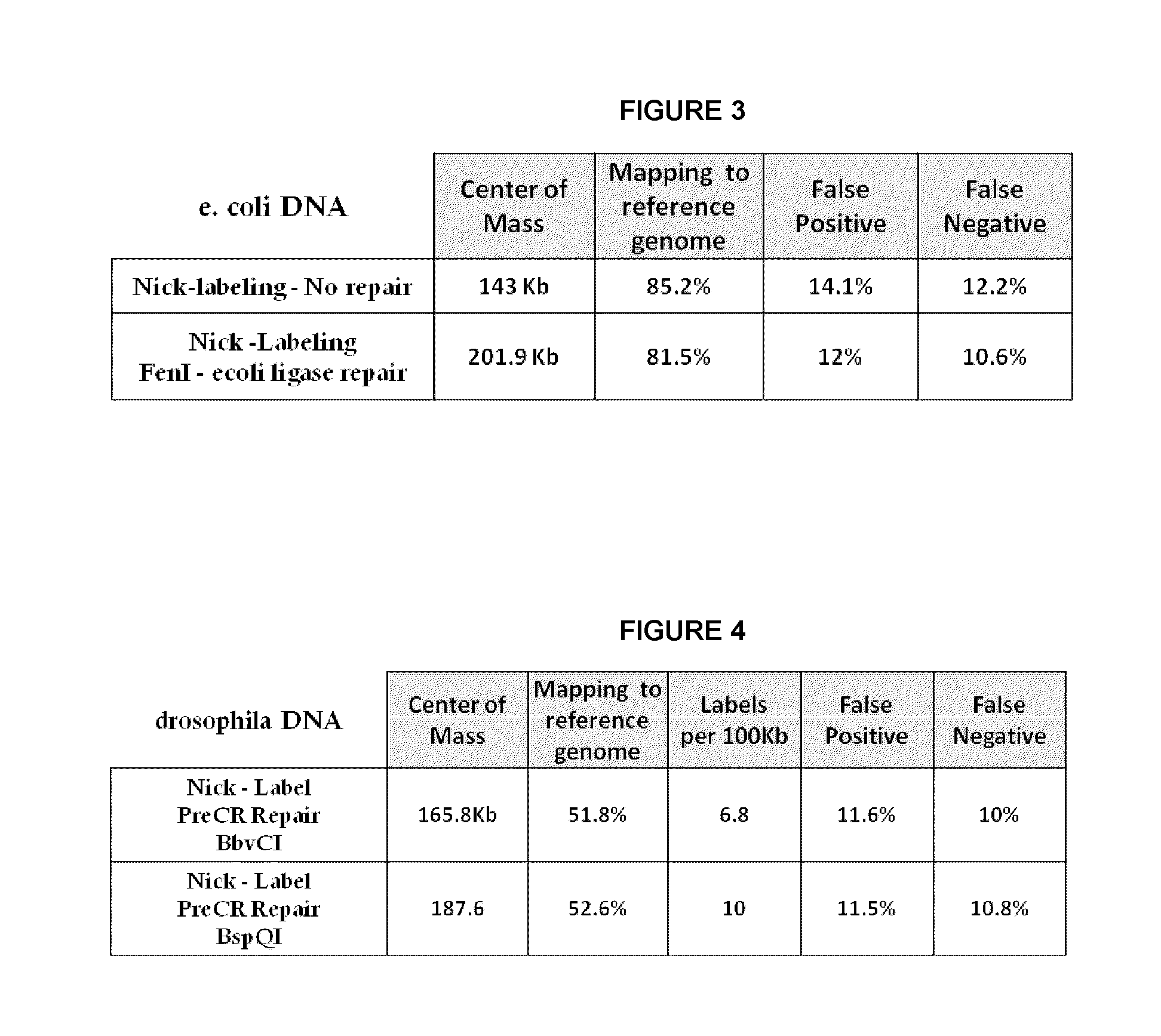
[0054]E. coli genomic DNA was nicked with Nt.BspQI nicking endonuclease. The nicked DNA was labeled with Taq polymerase by nick translation using Atto dUTP or Alexa dUTP in the presence of cold dATP, dGTP, and dCTP. The labeled nicks were: 1.) not repaired, 2.) repaired with PreCR as recommended by manufacture (New England BioLabs), 3.) repaired with PreCR under conditions of omitting dGTP, 4.) repaired with PreCR under conditions of omitting dATP and dGTP, or 5.) repaired with Taq polymerase under conditions of omitting dGTP. Ligation was then performed with a ligase. The resulting DNA was stained with YOYO-1 (Life Technologies) and processed on the Irys system (BioNano Genomics). Briefly, DNA was linearized in massively parallel nanochannels, excited with the appropriate laser for backbone and label detection, and optically imaged. Mapping to a reference genome, center of mass, and False Positive (FP) and False Negative (FN) calculations were carried out using nano Studio data ana...
example 2
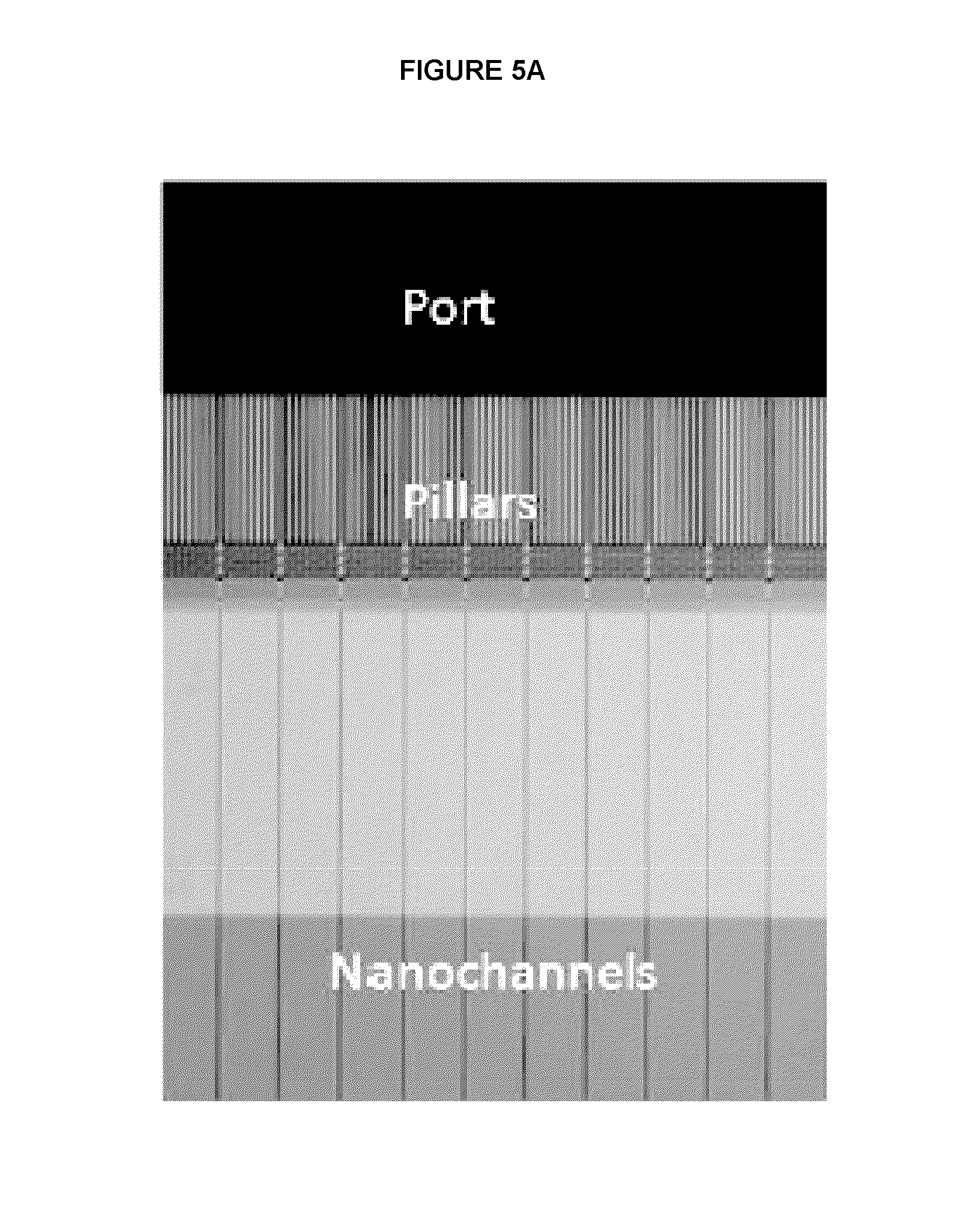
[0055]E. coli genomic DNA was nicked with Nt.BspQI nicking endonuclease. The nicked DNA was labeled with Taq polymerase by nick translation using Atto dUTP. The labeled DNA was: 1.) left unrepaired or 2.) treated with FEN Ito remove flaps followed by a ligase to repair the translated nicks. The DNA was linearized in massively parallel nanochannels, excited with the appropriate laser for backbone and label detection, and optically imaged. Mapping to a reference genome, center of mass, and False Positive (FP) and False Negative (FN) calculations were carried out using nano Studio data analysis software (BioNano Genomics). Results are shown in FIG. 3.
example 3
[0056]Drosophila genomic DNA was nicked with Nt.BspQI or Nb.BbVCI nicking endonuclease. The nicked DNA was labeled with Taq polymerase by nick translation using Atto dUTP. The labeled DNA was treated with PReCR reagent (New England Biolabs) to repair the nicks. The resulting DNA was stained with YOYO-1 (Life Technologies) and processed on the Irys system (BioNano Genomics). Mapping to a reference genome, center of mass, and False Positive (FP) and False Negative (FN) calculations were carried out using nano Studio data analysis software (BioNano Genomics). Results are shown in FIG. 4.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com