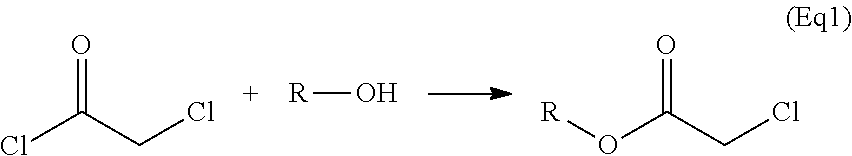
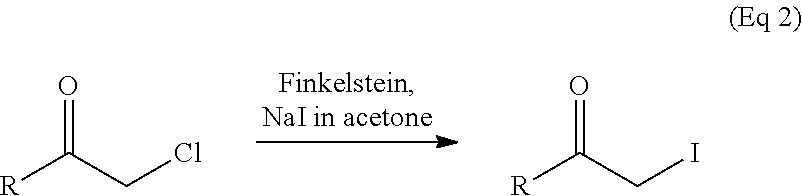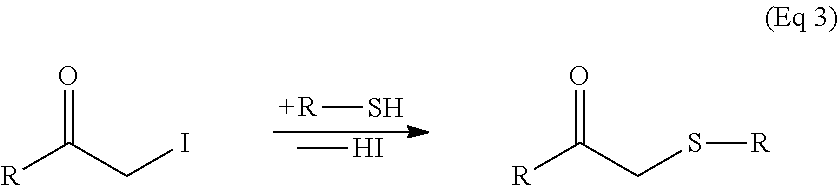Peptide libraries
a technology of peptides and libraries, applied in the field of peptides, can solve the problems of not recognising structure as well as structural diversity, and achieve the effects of increasing diversity, promoting even greater diversity, and reducing the number of structural isomers of peptide ligands
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Protease Resistant Bicyclic Peptide Against MDM2
[0251]MDM2 is an enzyme (an E3 ubiquitin ligase) that recognises the trans-activation domain of p53, the tumour suppressor, leading to ubiquitinylation and degradation of p53 by the proteasome. A nutlin inhibitor of the p53-MDM2 interaction can lead to in vivo activation of the p53 pathway, and it has been suggested that such agents may have potential as anti-cancer agents. Here we describe the selection of two bicyclic peptides (PEP10 and PEP48) against MDM2, a target “antigen”. The affinity of each synthetic peptide was in the range 250-750 nM.
[0252]Protocols generally followed those described earlier in Heinis et al., 2009, Nature Chemical Biology 5, 502-507, unless otherwise indicated. In the work of Heinis et al., both targets, kallikrein and cathepsin G, were proteases, and the kallikrein inhibitor is fairly resistant to proteolysis by kallikrein, although it includes a kallikrein cleavage site. MDM2 is not a protease, and theref...
example 2
Oxidation of PK15-TBMB Conjugate to its Sulfoxide and Sulfone
[0305]PK15-TBMB was synthesised as described in Heinis et al., 2009, Nature Chemical Biology 5, 502-507. Approximately 1 mg of PK15-TBMB was dissolved in 1 ml of 1×PBS and hydrogen peroxide added to a concentration of 0.3%. Reaction left at room temperature overnight. MALDI mass spec showed a range of peaks corresponding to the addition of 1,2 and 3 oxygen atoms, but incomplete reaction. Reaction adjusted to 1% H2O2 and left for 8 hrs where it could be seen that the +3 (oxygen) product was major but the reaction still incomplete. The reaction was heated in a microwave synthesiser, initially cooled on ice, up to a temperature of 37° C., with a power of up to 50 W. After 15 mins mass spec showed essentially a single peak at 1992 corresponding to addition of 3 oxygen atoms, and therefore corresponding to the sulphoxides. There is virtually no sign of addition of further oxygen atoms under these conditions. HPLC showed essenti...
example 3
Use of Trimethylmesitylene and Triethylmesitylene Cores
[0312]PK15 was conjugated with tris(bromomethyl)mesitylene under similar conditions to those described earlier with tris(bromomethyl)benzene (Heinis et al., 2009), and purified by HPLC. The conjugate was compared for its ability to inhibit kallikrein according to the procedure set forth in Example 1. Under conditions where PK15-TBMB inhibited kallikrein with an 1050 of 13 nM, preliminary results indicated that the hexamethyl benzene conjugate inhibited kallikrein with an 1050 of 150 nM. PK15 was also conjugated with tris(bromoethyl)mesitylene as above; this led to a further loss of inhibition, with an 1050 of about 400 nM. Thus the change in the nature of the core had a dramatic effect on the affinity of the ligand. In this case, the extra bulk of the three methyl or ethyl groups will likely have altered the packing around the core, and therefore the binding affinity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com