Nucleic acid hybrid film strip and reagent box used for diagnosing beta mediterranean sea thalassemia
A technology of thalassemia and nucleic acid hybridization, which is applied in the field of nucleic acid hybridization membrane strips for diagnosing thalassemia, and can solve problems such as difficult to distinguish, difficult to judge, and poor signal uniformity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
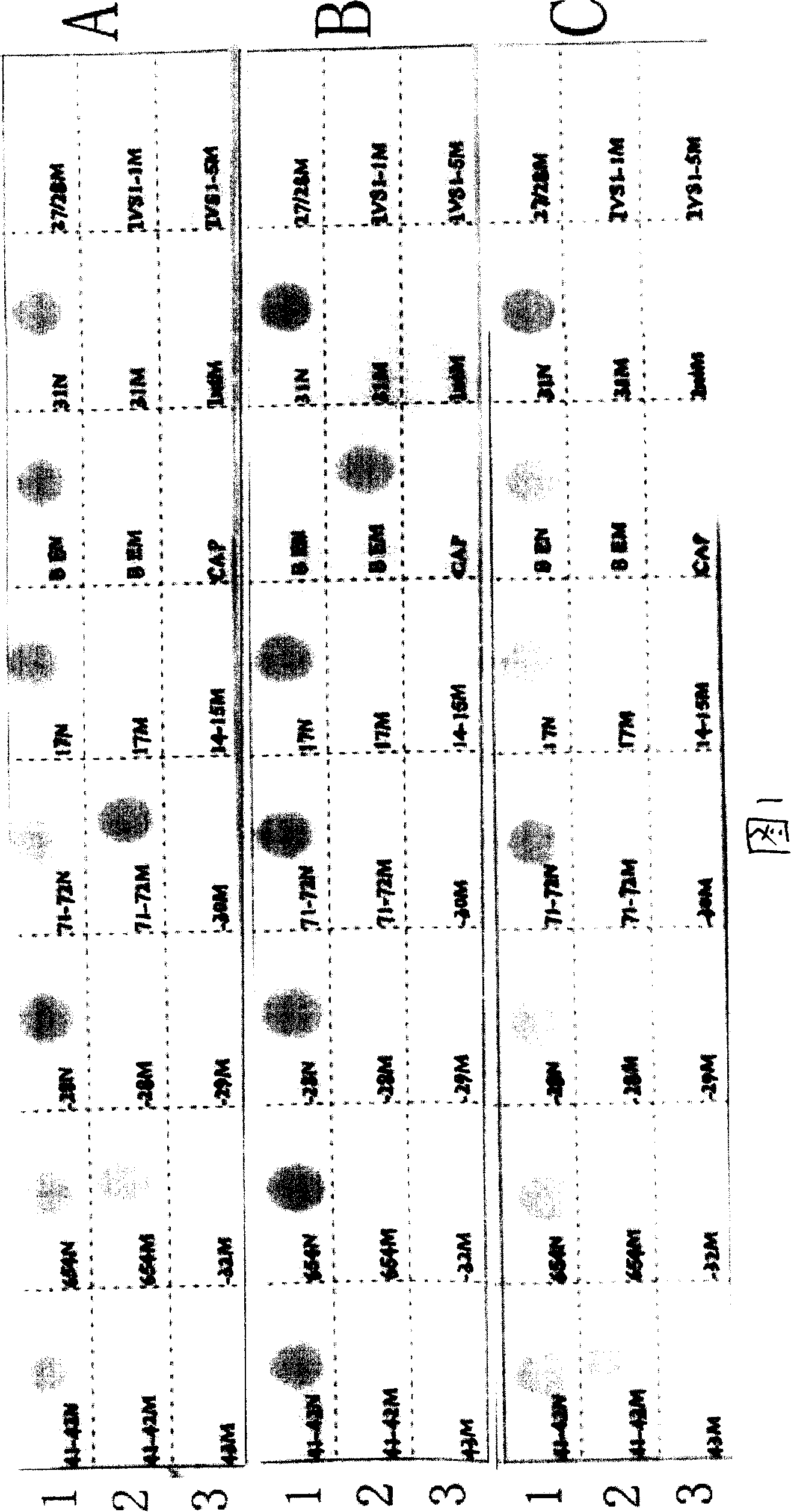
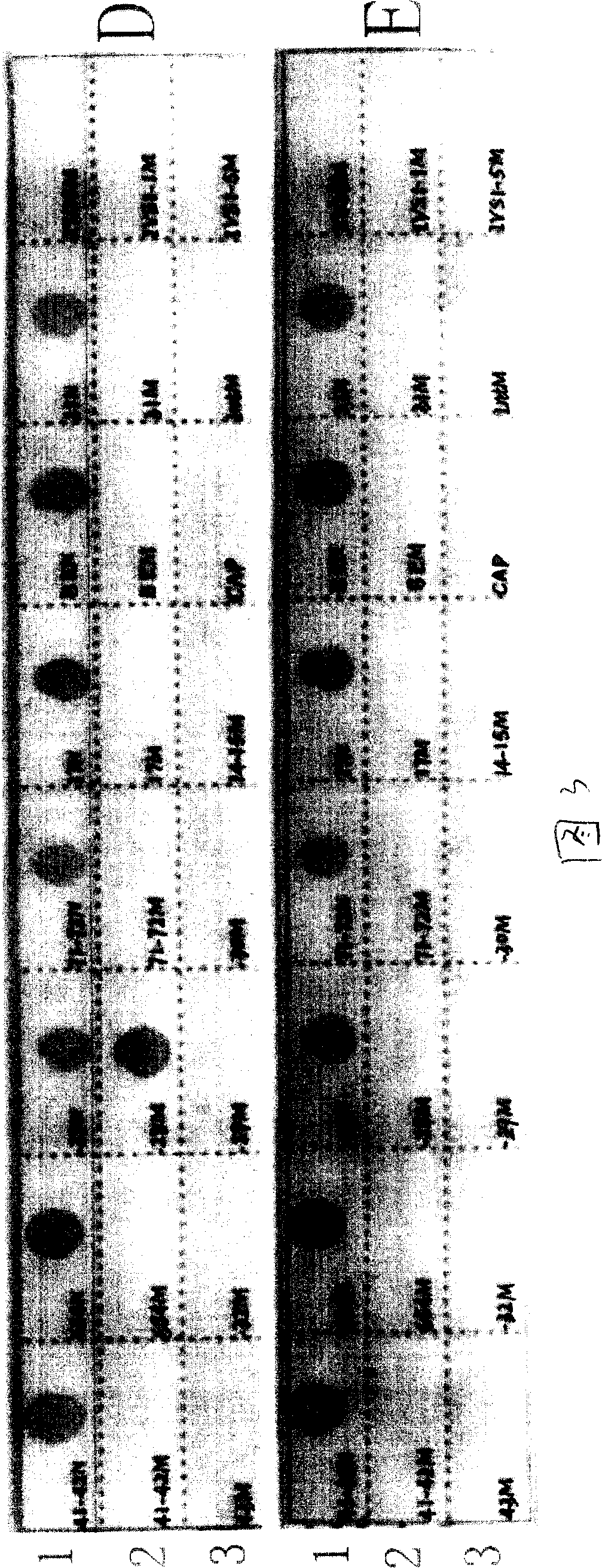
[0079] [Example 1] Preparation of nucleic acid hybridization membrane strips for detecting β-thalassaemia
[0080] 1. Preparation of probes
[0081] Probes were synthesized according to the sequences in Table 3, and the probes with reverse complementary sequences had the same effect as the sequences in the table, and will not be listed one by one in this table. Carrying out amino labeling on the 5' end or 3' end of the probe has the same effect, and the synthesis method is a conventional DNA synthesis method.
[0082] Table 3 DNA sequence of mutation probe
[0083] serial number
[0084] Note: W=A / T in the sequence of the probe IVS1-1M of SEQ ID NO.9.
[0085] Table 5 DNA sequence of normal probe
[0086] serial number
[0087] 2. Fabrication of nucleic acid hybridization strips
[0088] After the synthesis of oligonucleotide probes is completed, direct oligonucleotide probe immobilization is required. First cut the nylon membrane into a certain size, ...
Embodiment 2
[0092] [Example 2] Design of primers for amplifying the DNA in the sample to be tested
[0093] According to the sequence data of the β-thalassemia gene mutation, two pairs of specific primers with different lengths and Tm values and containing a biotin label at the 5' end were used to amplify two fragments of the β-globin gene. The nucleotide sequences of these two pairs of primers are shown in Table 6. In this table, the amplified products of SEQ ID NO.27 and SEQ ID NO.28 can be hybridized with 654N and / or 654M for detecting the mutation at position 654; the amplification of SEQ ID NO.25 and SEQ ID NO.26 The product was used to detect mutations at other sites except 654.
[0094] Table 6 DNA sequences of β-globin gene PCR primers of different lengths
[0095] serial number
Embodiment 3
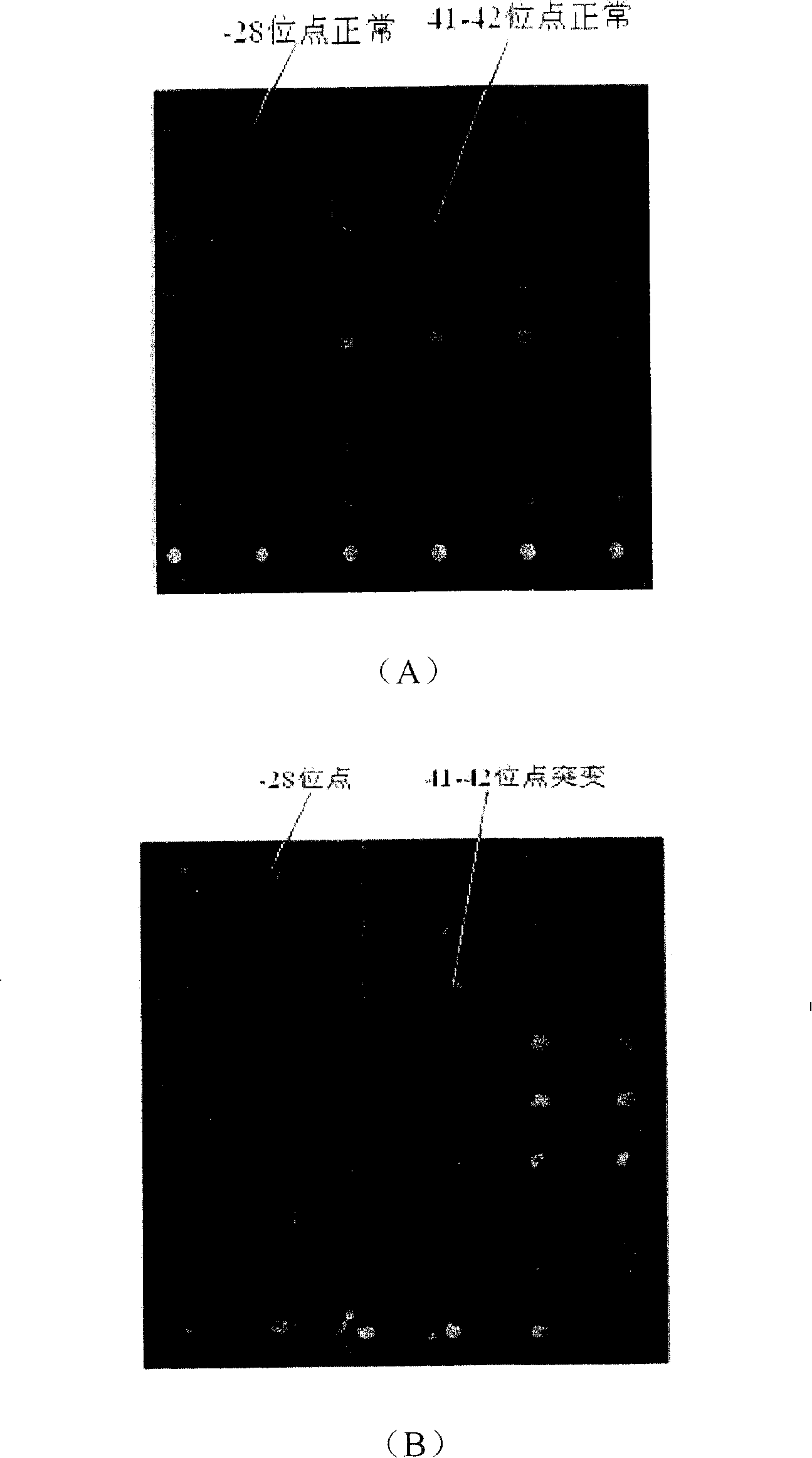
[0096] [Example 3] Detection of sample DNA to be tested
[0097] 1. Experimental instruments and reagents
[0098] 1. Test equipment
[0099] PCR instrument, DNA electrophoresis instrument, molecular hybridization box, shaker
[0100] 2. Test reagents
[0101] (1) 3% H 2 o 2
[0102] (2) 20×SSC (pH7.0): take 175.3g of NaCl, 88.2g of sodium citrate, add H 2 Dissolve O to 1000 mL, adjust the pH to 7.0 with a pH meter, and autoclave.
[0103] (3) 10% SDS (pH 7.0): Dissolve 20g of SDS in 180mL of H2O, adjust the pH to 7.0 with HCl, and finally set the volume to 200mL.
[0104] (4) Liquid A (2×SSC, 0.1% SDS, pH 7.4): take 100mL of 20×SSC, 10mL of 10% SDS, add H 2 O was dissolved to 1000mL, and the pH was adjusted to 7.4.
[0105] (5) Solution B (0.5×SSC, 0.1% SDS, pH 7.4): take 20×SSC 25mL, 10% SDS 10mL, add H 2 O was dissolved to 1000mL, and the pH was adjusted to 7.4.
[0106] (6) Liquid C (0.1M sodium citrate, pH 5.4): Dissolve 14.7 g of sodium citrate in 450 mL of wate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com