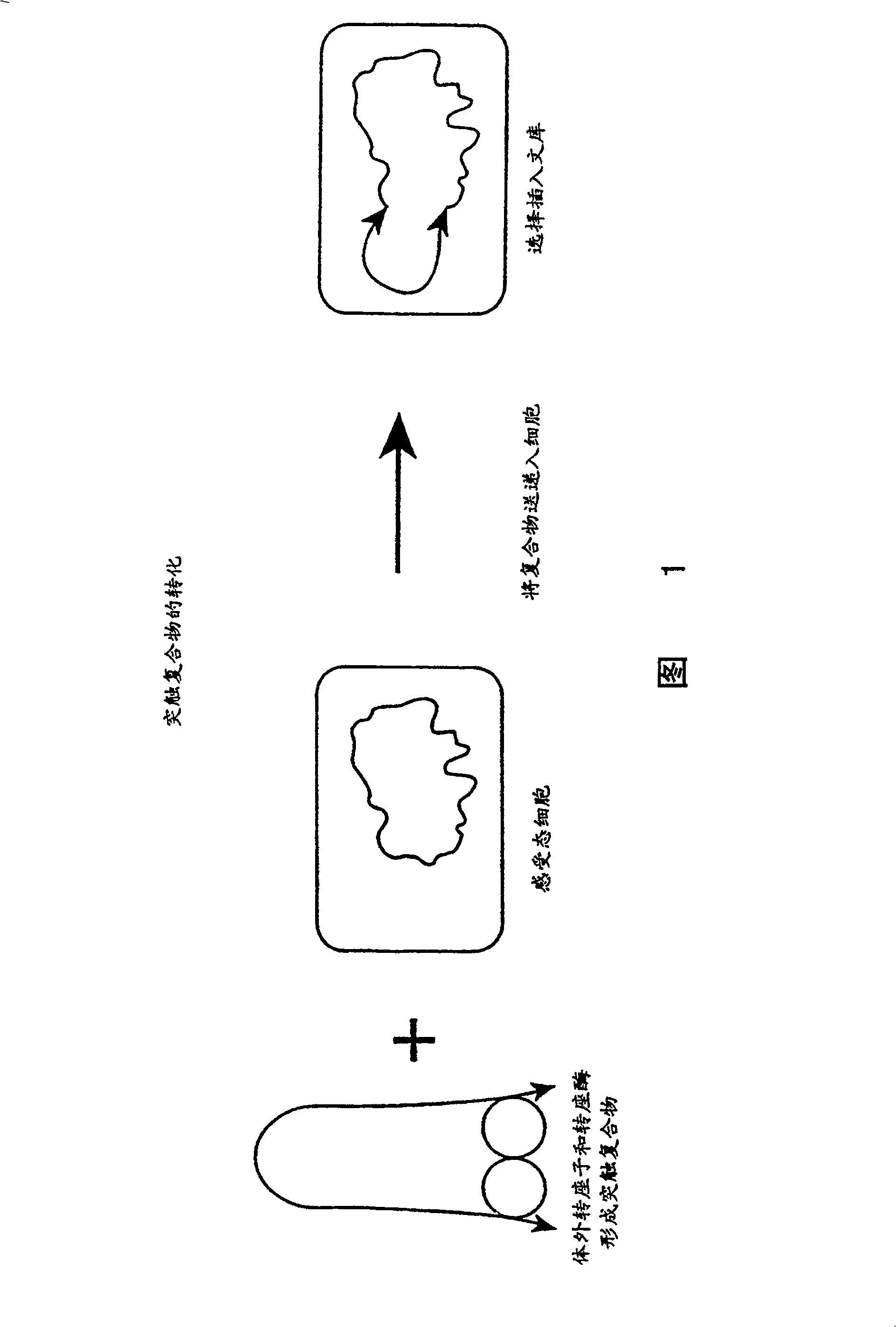
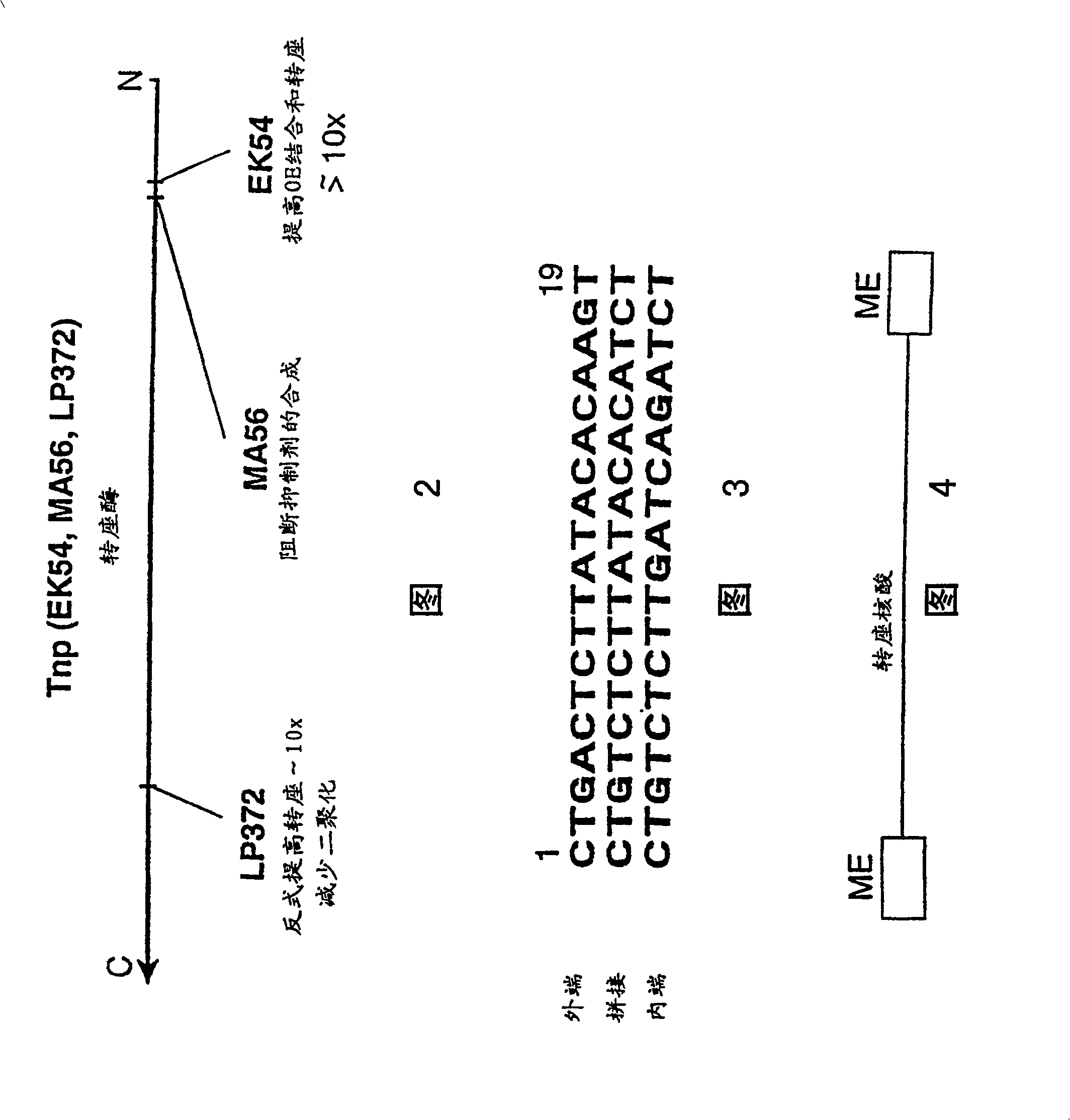
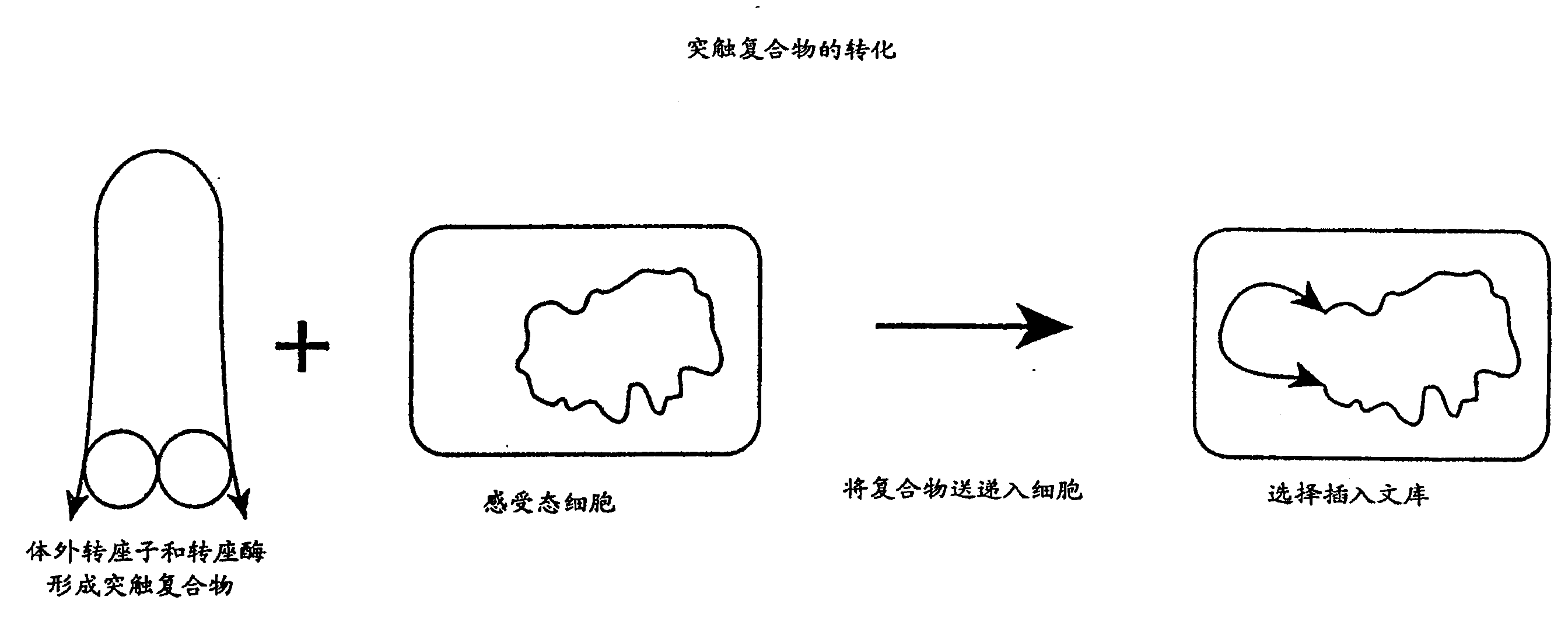
Method for making insertional mutations
A technology of insertion mutation and synapse, which can be applied to other methods of inserting foreign genetic materials, mutant preparation, botanical equipment and methods, etc., and can solve the problems of complicated and unsatisfactory insertion mutation library analysis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0035] In a 1 μl, 0.05 μg purified hyperactive transposase (EK54 / MA56 / LP372) reaction, the amino acid sequence disclosed in the international application PCT / US97 / 15941 cited herein was combined with 0.1 μg transposable polynucleotide, the transposable polynucleotide The locus polynucleotide contains an expression cassette encoding a protein that confers kanamycin resistance to target cells. This expression cassette is flanked by spliced ends (as described in the same international application cited) and is shown in Figure 3. Separated The polynucleotide was provided in a reaction in , either as a supercoiled plasmid, a linearized plasmid, or as a fragment of the polynucleotide including the reverse sequence required for Tn5 transposase-mediated transposition at its terminus.
[0036] With or without magnesium ions (Mg ++ ) in reaction buffer for 1 hour. In the absence of magnesium ions, synaptic complexes formed in vitro but no transposition occurred.
[0037] After incuba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com