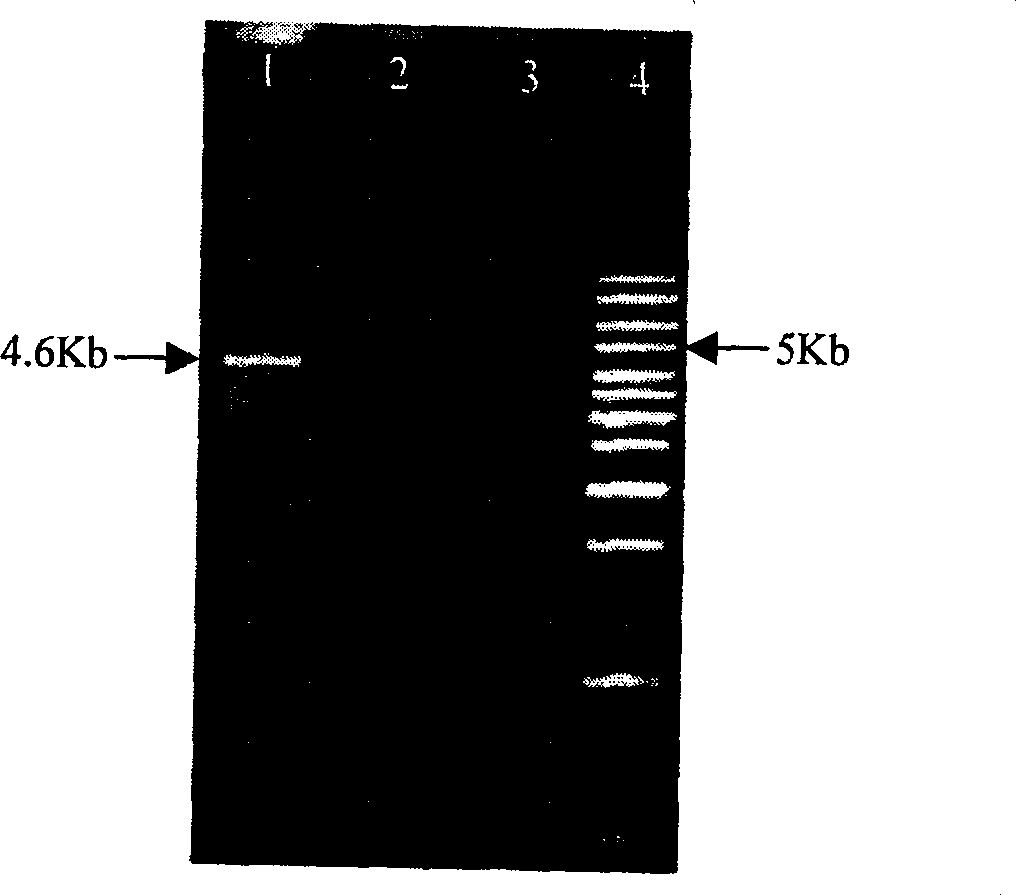
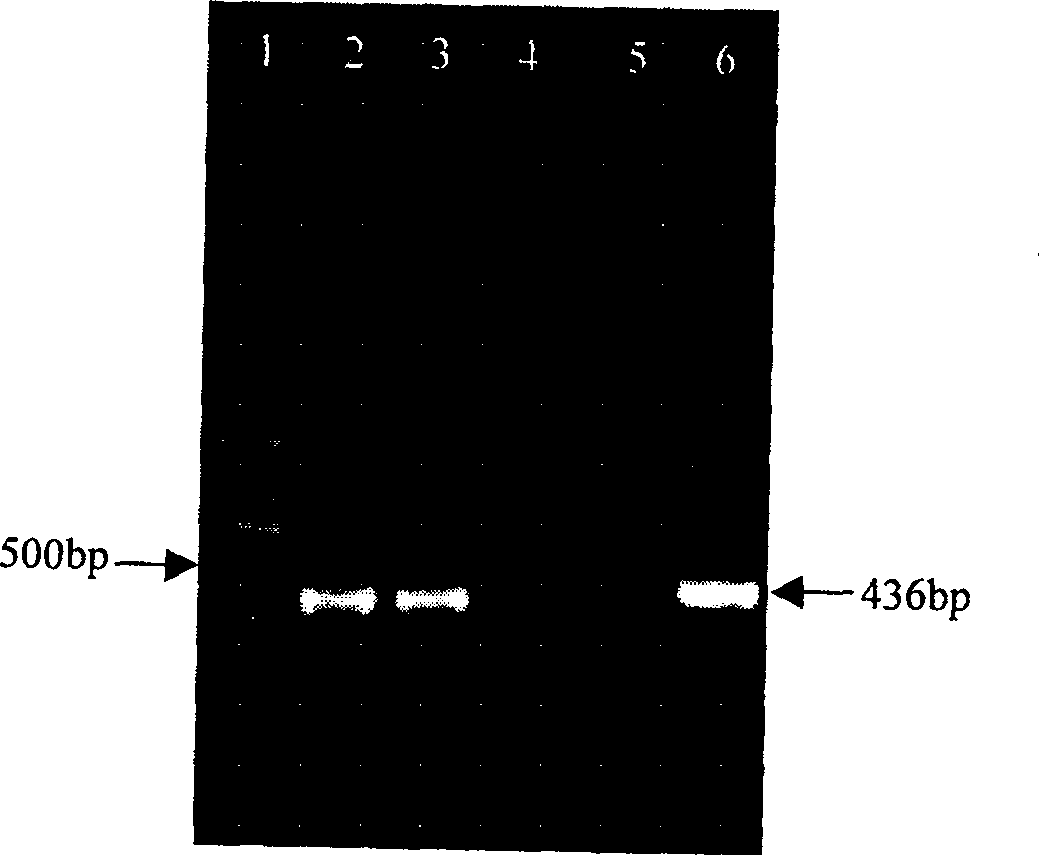
Bovine genome pseudo-attP site and its use
A genome and gene technology, applied in the fields of application, genetic engineering, plant genetic improvement, etc., to achieve the effect of promoting further application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 , Construction of plasmid pCMVInt containing Streptomyces phage φC31 integrase gene
[0028] According to the method described in the literature (Groth A.C., Olivares E.C., Thyagarajan B.and Calos M.P..Proc.Natl Acad.Sci.USA 97(2000):5995-6000), construct the plasmid pCMVInt containing the integrase gene of Streptomyces phage φC31 , the construction is divided into two steps, as follows:
[0029] The first step, the construction of plasmid pTA-Int:
[0030](1) Genomic DNA of Streptomyces phage φC31 (German Collection Center for Microbiology and Cell Culture, Braunschweig, Germany, DSM No. 49156) was used as a template to amplify the integrase gene by PCR.
[0031] The primer sequences are:
[0032] 5'-GAGTTCTCTCAGTACTAGTCGTAGGGTCGC-3'; and
[0033] 5'-CGCCTAACAGGGGATCCGGTGTCTCGCTAC-3';
[0034] PCR reaction system: 1×PCR buffer solution (TaKaRa company), 1.5mM Mg 2+ , 10 μM each primer, 200 μM dNTPs, 100-200 ng template DNA, 2.5 U LATaq DNA polymerase (...
Embodiment 2
[0047] Example 2 , construction of plasmid vector containing attB site
[0048] According to the method described in the literature (Groth A.C., Olivares E.C., Thyagarajan B.and Calos M.P..Proc.Natl Acad.Sci.USA 97(2000):5995-6000), construct the plasmid pBCPB+ containing the attB site, the specific process is as follows :
[0049] The first step, the construction of plasmid pTA-attB:
[0050] (1) With a pair of primers:
[0051] 5'-CAGGTACCGTCGACGATGTAGGTCACGGTC-3'; and
[0052] 5'-GTCGACATGCCCGCCGTGACCG-3'
[0053] Using the genomic DNA of Streptomyces lividans (German Microbiology and Cell Culture Collection Center, Braunschweig, Germany, DSM No.46482) as a template, a 293bp sequence was amplified by PCR, which contained the φC31 integrase recognition attB site.
[0054] PCR reaction system: 1×PCR buffer solution (TaKaRa company), 1.5mM Mg 2+ , 10 μM each primer, 200 μM dNTPs, 100-400 ng template DNA, 2U Taq DNA polymerase (TaKaRa Company).
[0055] PCR reaction co...
Embodiment 3
[0099] Example 3 , construction of a plasmid containing the gene of interest (EGFP) and attB site
[0100] In this example, unless otherwise specified, all restriction endonucleases were purchased from TaKaRa Company.
[0101] The construction of plasmid pEGFP-N1-attB is divided into three steps:
[0102] Step 1: Construction of plasmid pSL301-attB
[0103] (1) Restriction plasmid pBCPB: pBCPB obtained in Example 2, 25 μl; Xho I, 4 μl; 10×H, 20 μl; sterilized double distilled water, 151 μl; total reaction volume, 200 μl. 37°C overnight.
[0104] (2) Restriction plasmid pSL301: pSL301 (Invitrogen), 25 μl; Xho I, 4 μl; 10×H, 20 μl; sterilized double distilled water, 151 μl; total reaction volume 200 μl. 37°C overnight.
[0105] (3) Dephosphorylation reaction of the linearized fragment of plasmid pSL301: Precipitate the digested product of pSL301 with 2 times the volume of absolute ethanol, wash once with 70% ethanol, dissolve the linearized vector in 43 μl sterile double d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com