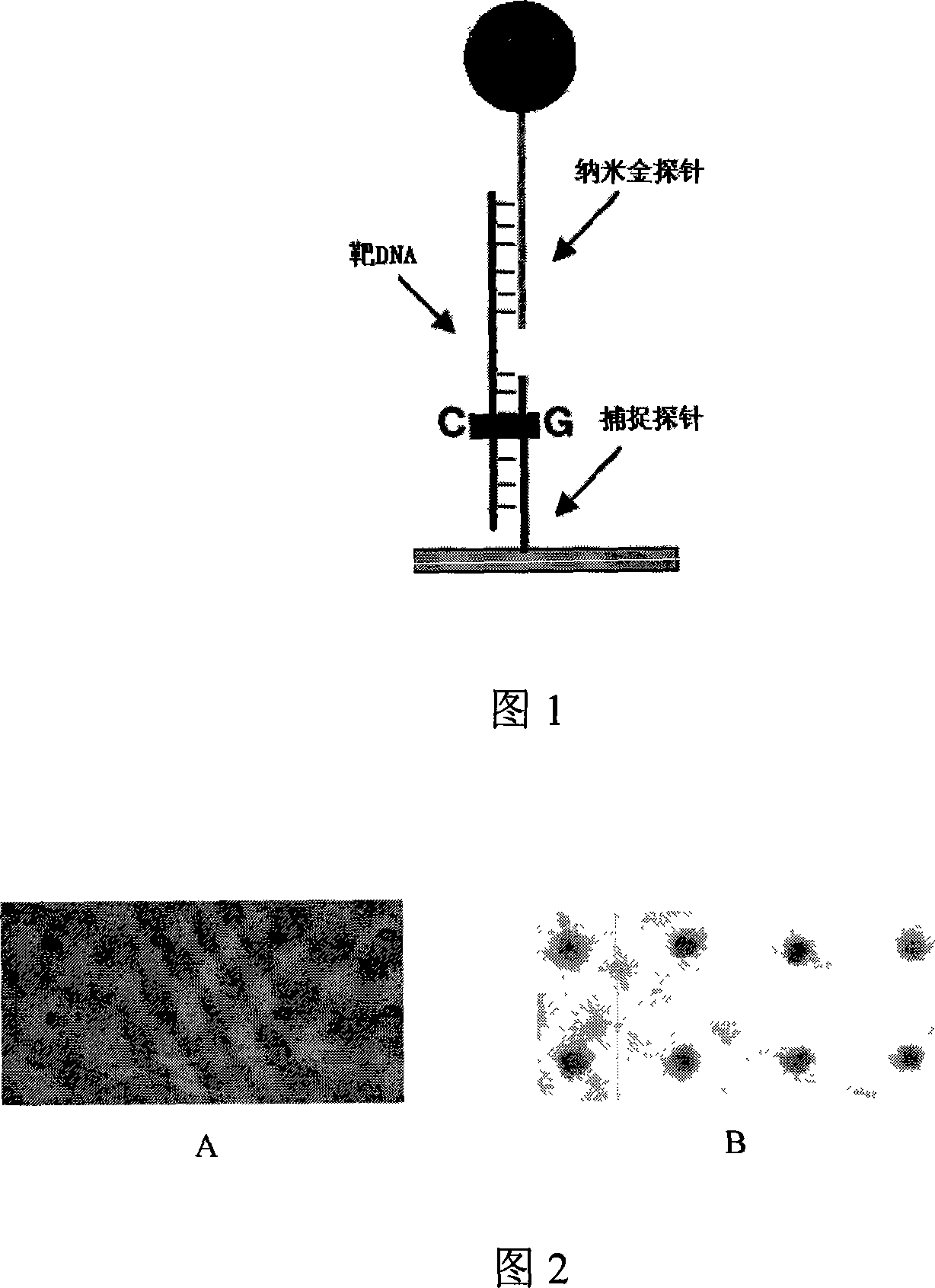
Nanometer detecting probe chip without amplifying genom DNA and detection method
A technology of nano-probe and detection method, which is applied in the field of nano-probe chip to achieve the effects of convenient popularization, good repeatability and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0036] The present invention will be further described below in conjunction with the embodiments of the present invention and the accompanying drawings.
[0037] Example Nanoprobe chip for P53 gene detection
[0038] 1. Prepare materials and reagents
[0039] (1) Chip: self-made;
[0040] (2) EasyHtyb hybridization solution: purchased from Roche Company;
[0041] (3) All probes were synthesized in Dalian Bao Biological Engineering (Dalian) Co., Ltd. (Dalian TaKaRa Company)
[0042] (4) 15nm and 30nm gold nanoparticles were purchased from Shanghai Water Source Biotechnology Co., Ltd.
[0043] (5)NaNO 3 Washing solution: 1M NaNO 3 300ml
[0044] 0.2M Na 2 HPO 4 30.5ml
[0045] 0.2M NaH 2 PO 4 9.5ml
[0046] Dilute to 1000ml
[0047] (6) PB solution: 0.3mol / L NaNO 3 and 10mmol / L Na 2 HPO 4 / NaH 2 PO 4 , PH7.0
[0048] (7) Silver stain enhancement solution: 60 μl of 1% gelatin
[0049] 2.55g citri...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com