Method for separating short interspersed repeated segments based on magnetic bead probe complexes
A repetitive sequence and compound technology, applied in recombinant DNA technology, DNA preparation, etc., can solve the problems of heavy workload, high cost, and low efficiency, and achieve the effects of reducing cost consumption, easy control, and improving experimental efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
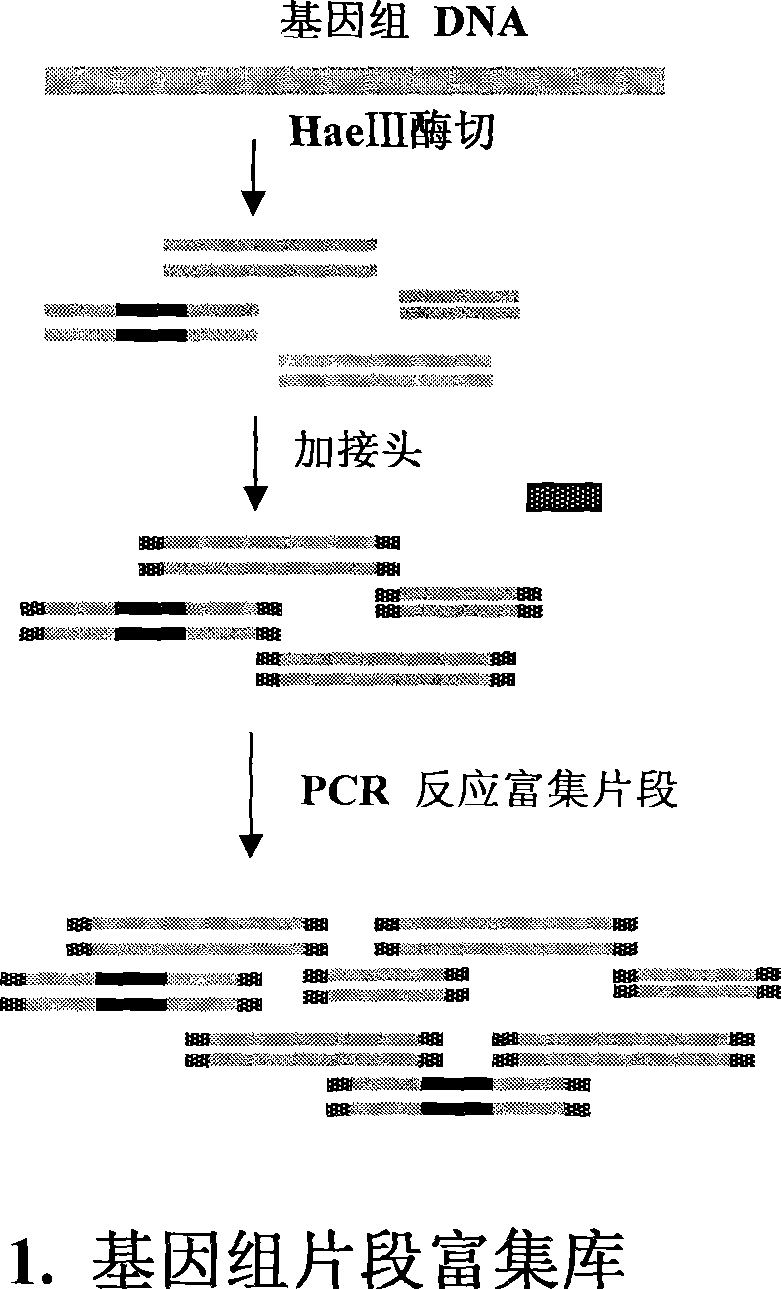
[0024] Example 1: The whole process is illustrated in the case of bighead carp.
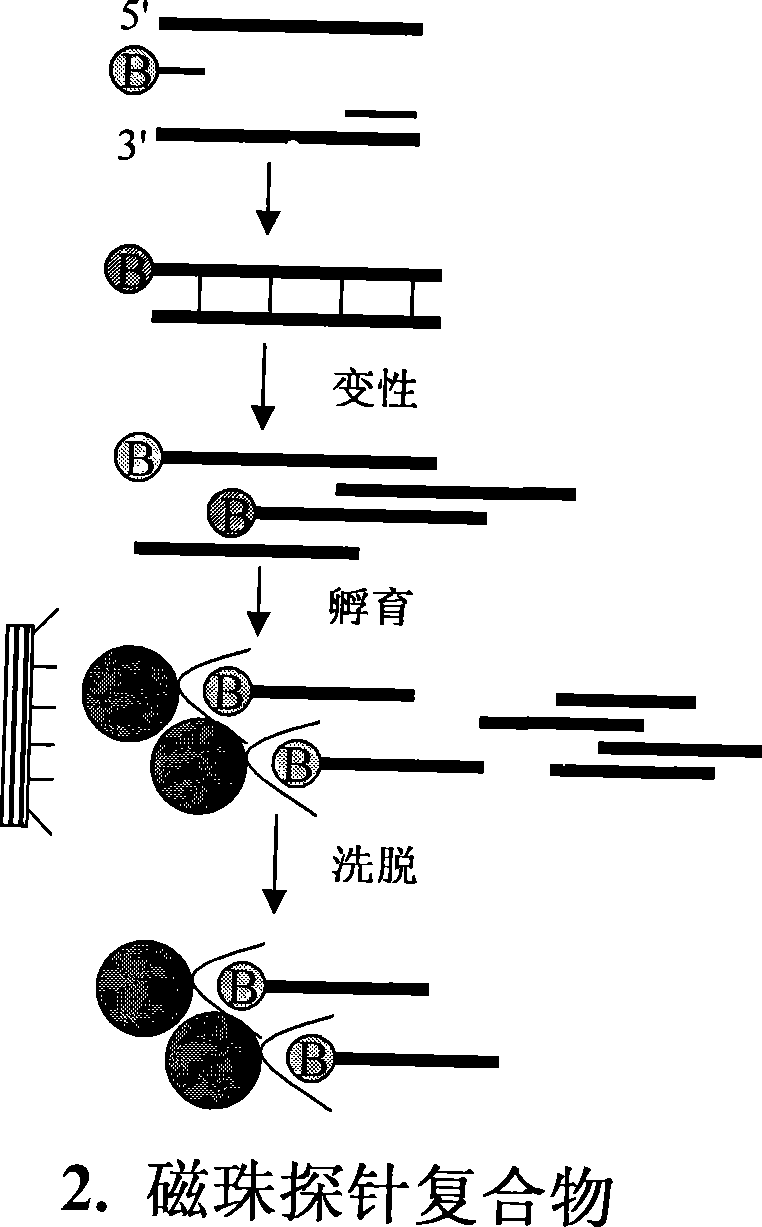
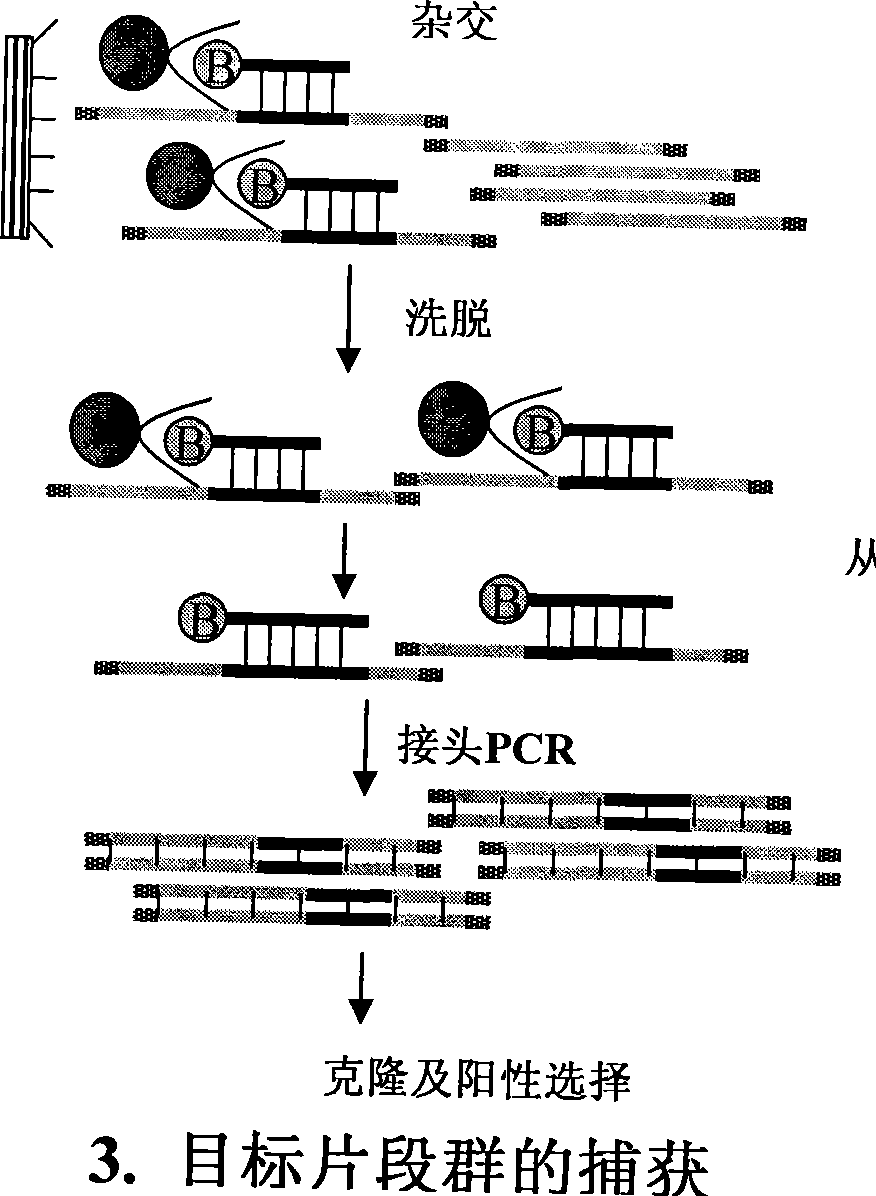
[0025] A. According to figure 1 It can be seen that the preparation of the genome fragment enrichment library. Step 1: Enzyme digestion of genomic DNA. About 40 micrograms of genomic DNA and 20 units of restriction endonuclease HaeIII were added to 100 microliters of enzyme digestion system, and digested overnight for complete digestion. The map after digestion is as figure 2A, Genomic DNA was uniformly digested below 4KB, and the recovered 500bp-2kb fragment was dissolved in 40 microliters of sterile water. The second step: add joints. The two oligonucleotides that generate the linker are A: 5P'GGCAGGATCCACTGAATTCGC-3' and B: 5'-AGCGAATTCAGTGGATCCTGCC-3'. The two oligonucleotides were carried out in sterile water with a final concentration of 10 μM to perform the following procedures to achieve the polymerization of the two strands to form a linker: 95°C for 3 minutes, 65°C for 2 minutes, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com