Prevention/treatment of ichthyosis vulgaris, atopy and other disorders
An ichthyosis, common technology, used in gene therapy, microbial assay/test, skin diseases, etc., can solve problems such as unidentified genomic mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Co-reporter gene constructs for testing read-through reagents
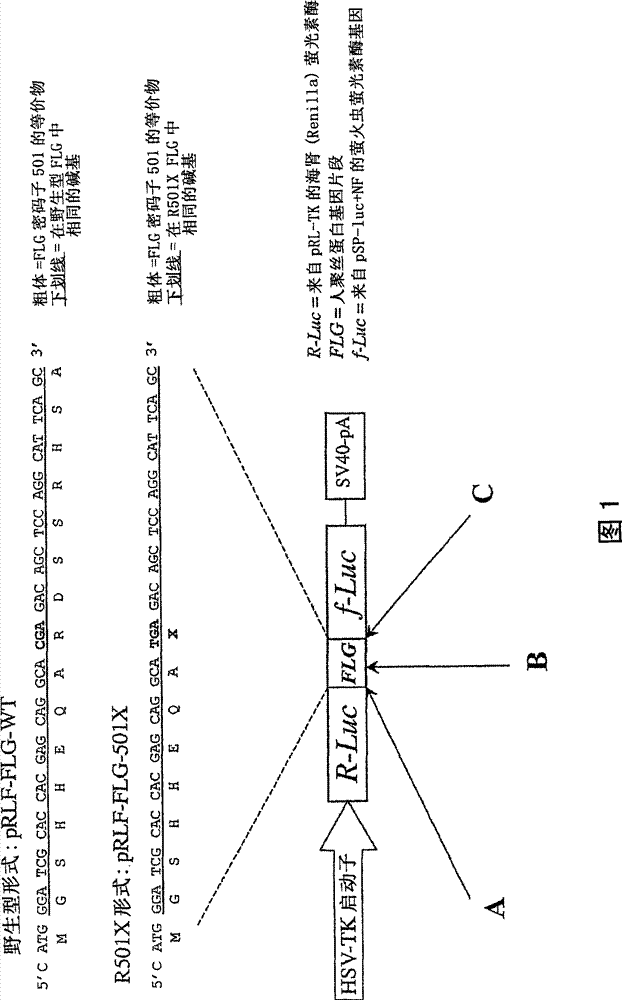
[0044] The oligonucleotide cassette corresponding to base numbers 1480-1523 of the human filaggrin gene, (FLG; coding sequence numbered starting from the ATG start codon; Genbank accession number NM_002016.1), was cloned into the unique Nco I restriction This site spans the ATG codon of the firefly luciferase gene (f-Luc) in the plasmid vector pSP-luc+NF (Promega). figure 2 The oligonucleotide cassette shown in has an added overhang corresponding to the cohesive end of Nco I. These boxes correspond to the region of the human FLG gene containing codon 501, the common R501X mutation site. Wild type and R501X mutant forms were formed. Clones with inserts in the correct orientation were identified and verified by DNA sequencing. After insertion of the FLG cassette, the ATG codon, the original start codon of the f-Luc gene, was mutated to an AGG arginine codon by site-directed mutagenesis using th...
Embodiment 2
[0047] Example 2: Generation of Human Suppressor tRNA Genes for TGA Nonsense Mutations
[0048] The human genome includes a large number of CGA-arginine t-RNA genes. These are very compact genes and contain an intragenic promoter for RNA polymerase III (Lewin B., In: Genes VII, Oxford University Press, 2000, pp. 624-626). Two of these genes were identified in the current collection of the human genome (HG17; http: / / genome.ucsc.edu), named TR3 and TR4B for convenience, and amplified from normal human control DNA . TR3 is located on chromosome 6p22.1 and TR4B is located on chromosome 15q26.1. Specifically, using primers TR3.F5'CGA TGC AGA CAA TAT GCA GA3' and TR3.R5'CTAAGC CTA CAAAAC CGAAA3', the TR3 gene was amplified as a 668bp fragment, which corresponds to the base number in the human genome HG17 collection Chr6: 26,407,555-26,408,284. The TR4B gene was amplified as a 465 bp fragment using primers TR4B.F5'GAC TTC TGG GTG GGCTCT CC3' and TR4B.R5'CCC TCC CTC TCC ACT TTC CT...
Embodiment 3
[0049] Example 3: Dual Luciferase Assay for FLG Readthrough
[0050] The co-reporter constructs pRLF-FLG-WT and pRLF-FLG-501X were transiently transfected into 293 cells (transformed human kidney epithelial cell line) in 96-well plates by utilizing the Fugene-6 system (Roche) , and test them. Forty-eight hours after transfection, the cells were subjected to the Dual Luciferase Assay System (Promega), which detects specific luciferase signals corresponding to Renilla and Firefly luciferase reporters. For the wild-type co-reporter gene, pRLF-FLG-WT, a strong signal for Renilla and firefly luciferase was obtained, as shown in Figure 6 as shown in a. In contrast, only the Renilla signal was obtained for the pRLF-FLG-501X construct, showing that the R501X premature stop codon mutation present within the FLG box in this construct resulted in complete loss of firefly luciferase expression ( Figure 6 a).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com