PNA, probe, primer and method for detecting K-ras gene mutation
A probe and gene technology, applied in the field of PNA to detect K-ras gene mutation, can solve the problems of cumbersome steps and achieve the effect of simple operation, high sensitivity and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
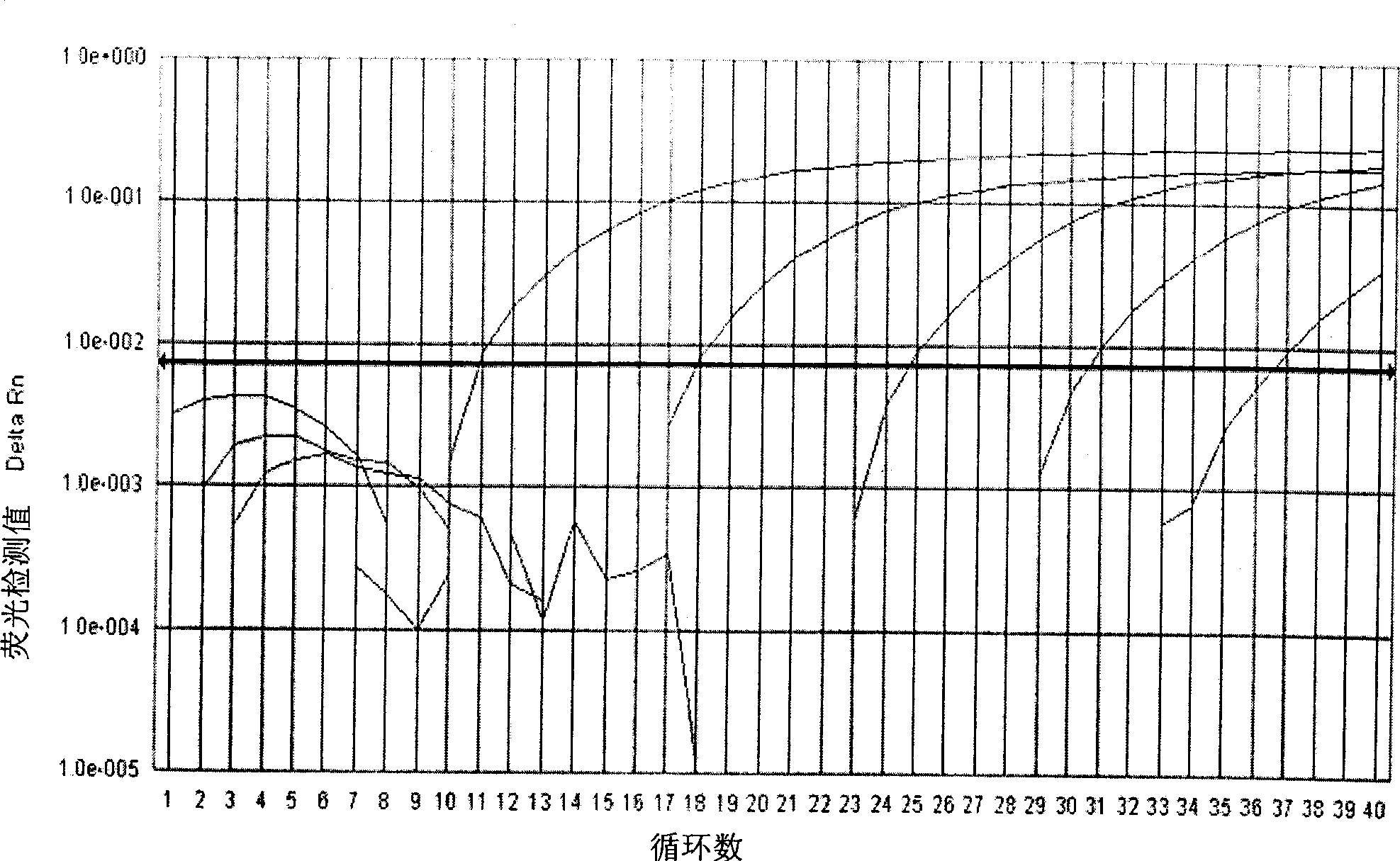
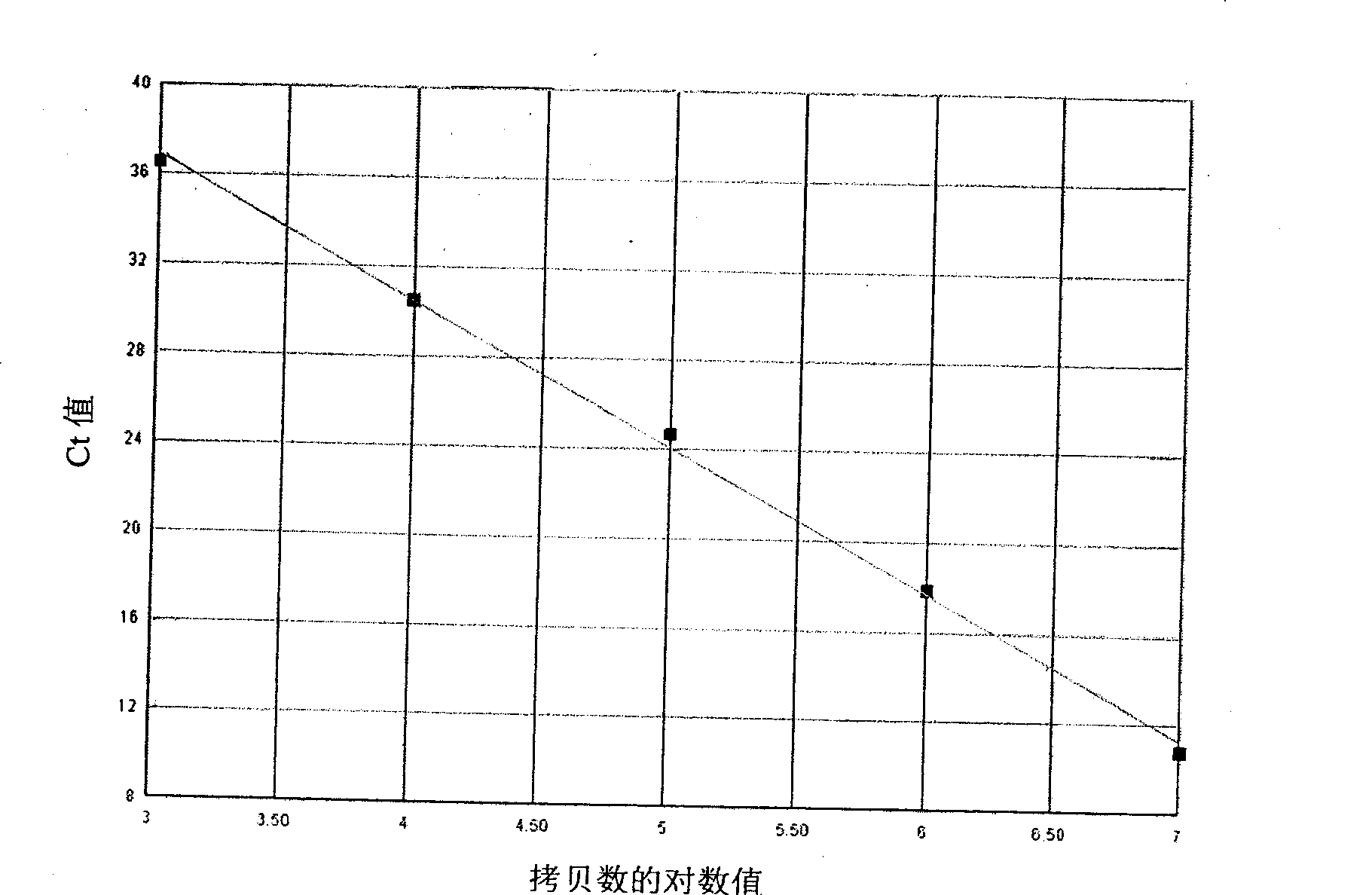
[0049] Example 1 Carrying out real-time quantitative PCR detection on the plasmid standard substance of known mutation amount
[0050] 1. Main source of reagents:
[0051] 1.1 Designing PNAs
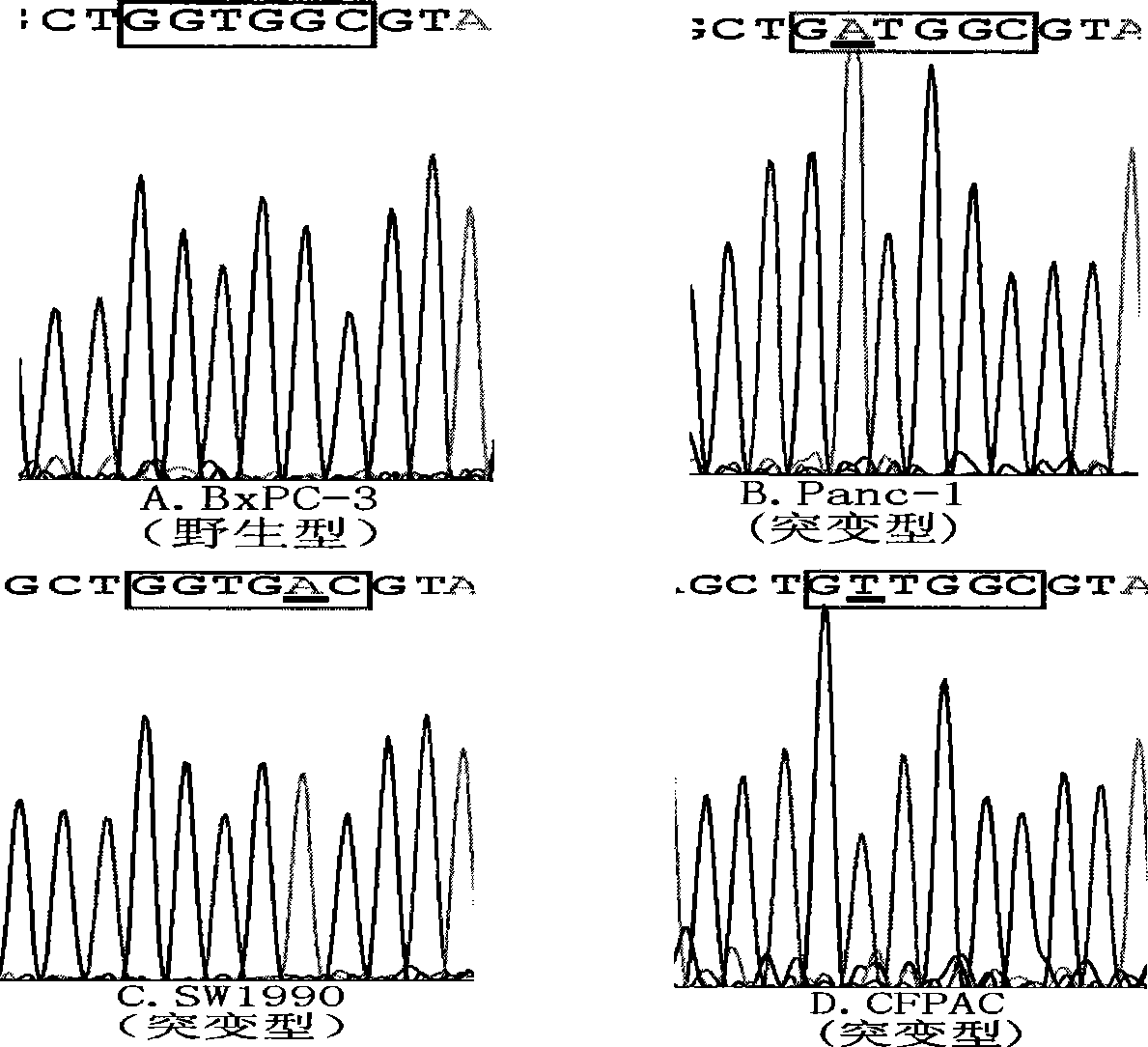
[0052] PNA is a PNA designed for the 12th and 13th codons of the wild-type K-ras gene, comprising at least 12 consecutive bases of SEQ ID NO: 1 or its complementary sequence, and the preferred sequence of the PNA is Ac-ACGCCACCAGCTC-Lysine-COOH (SEQ ID NO: 1) ID NO: 1), synthesized by South Korea panagene company.
[0053] 1.2 Design of Tagman MGB probe
[0054] The Tagman MGB probe hybridizes to the wild-type K-ras gene target sequence, the probe sequence comprising at least 15 consecutive nucleotides of SEQ ID NO: 2 or its complement. The probe is preferably an oligonucleotide sequence shown in SEQ ID NO: 2, i.e. FAM-AGCTCCAACTACCACAAG-MGB (SEQ ID NO: 2), and its two ends are respectively labeled with a reporter group FAM and a quencher group MGB. Needles were synthesized by Shangh...
Embodiment 2
[0105] Example 2 Detection of K-ras gene mutation in clinical samples (taking blood and stool samples as examples)
[0106] 1. Collect the feces and blood of patients with clinical pancreatic cancer, chronic pancreatitis and normal people, and extract DNA.
[0107] 2. Blood DNA extraction method: ① anticoagulated whole blood 0.5 ~ 12ml; ② add 500 ~ 700ulTT buffer, shake vigorously, centrifuge at 12000 rpm for 5 minutes in a low temperature centrifuge; ③ collect the precipitate, repeat steps 2 and 3 , until the precipitate is colorless; ④Add 200ul PCR lysate A; ⑤37 ℃ water bath overnight; Add 200ul of chloroform to the supernatant, mix thoroughly, and centrifuge at 12000 rpm for 10 minutes; / centrifuge for 10 minutes; ⑨discard the supernatant and add 1ml of 70% ethanol to wash, 12000 rpm / centrifuge for 5 minutes; ⑩discard the supernatant, air-dry naturally, dissolve the precipitate with 100ulTE solution, aliquot and store at -80°C.
[0108] 3. Fecal DNA extraction method: Qia...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com