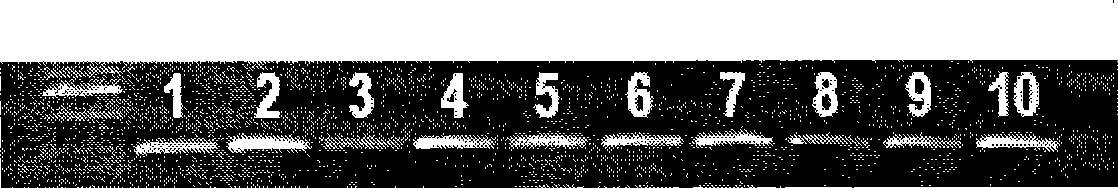Simple and new method for identifying copy numbers of transgenic plants
A technology of transgenic plants and copy number, applied in the field of molecular detection of transgenic plants, can solve the problems of expensive experimental equipment and consumables, and achieve the effects of increasing the difficulty of the experiment, shortening time-consuming and low-cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0040] Example 1. Determination of the copy number of the offspring of transgenic Bt gene insect-resistant cotton
[0041] 1 Routine PCR molecular detection.
[0042] Extract the total DNA of the transgenic Bt gene insect-resistant cotton sample, adjust the DNA concentration of the sample to 100ng / μl, and do PCR molecular detection. figure 1 It is the agarose gel electrophoresis picture of the PCR products of 10 transgenic materials. Among the positive reactions, the PCR product band of the No. 2 individual strain is the brightest, and the DNA of the No. 2 individual strain is used to determine the cycle number.
[0043] 2 Detection of target gene primers of transgenic Bt cotton and primers of internal standard single-copy genes
[0044] Primer sequences of Bt gene:
[0045] cry1A-F: 5'GAAGGATTGAGCAATCTCTAC 3'
[0046] cry1A-R: 5'CAATCAGCCTAGTAAGGTCGT 3'
[0047] The expected amplified fragment size is 340bp.
[0048] Internal standard single copy gene SAD1 primer sequenc...
example 2
[0062] Example 2. Determination of the copy number of the offspring of transgenic Susy cotton
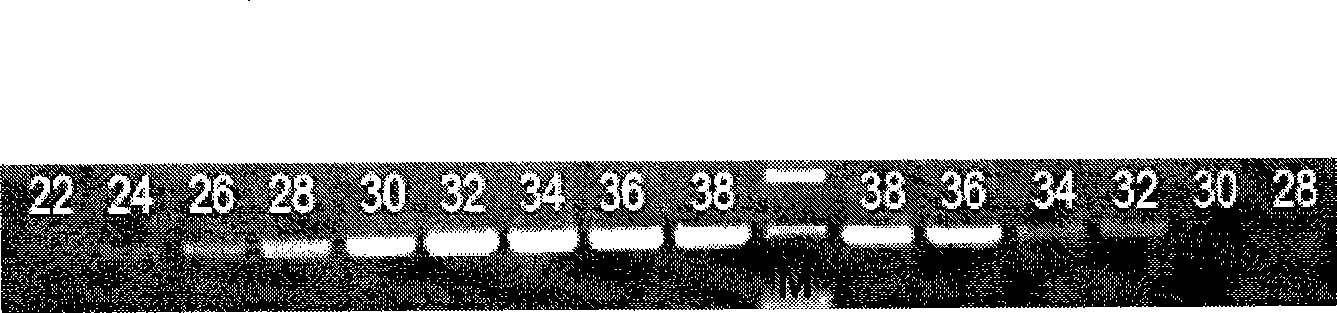
[0063] 1. According to the experimental process of Example 1, the optimal number of PCR cycles to determine the transgenic Susy cotton is 32 cycles.
[0064] 2. Target gene primers
[0065] Susy-F: 5'CATCATTGCGATAAAGGAAAGG 3'
[0066] Susy-R: 5'GGCTCAAAATCCAATTCCAAAAA 3'
[0067] The expected amplified fragment size is a fragment of 720bp.
[0068] 3. Determination of copy number
[0069] Divide the PCR reaction mixture of the sample to be tested (20μl PCR system, 2.0μl 10×Taq Buffer, 1.6μl dNTP (2.5mM), 0.2μl Taq enzyme, 14.6μl water, 1.0μl DNA template) into 2 equal parts One part was added to the target gene Susy primer, and one part was added to the internal standard gene SAD1 primer. The target Susy gene cycling conditions were 94°C preheating for 2 minutes, 94°C for 30 s, 58°C for 30 s, 72°C for 1 min, and 32 cycles of 72°C extension for 10 min. The internal standard SAD...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com