Patents
Literature
61 results about "Single copy gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Single copy DNA (scDNA) nucleotide sequences present once in the haploid genome, as are most of those encoding polypeptides in the eukaryotic genome. spacer DNA the nucleotide sequences occurring between genes.
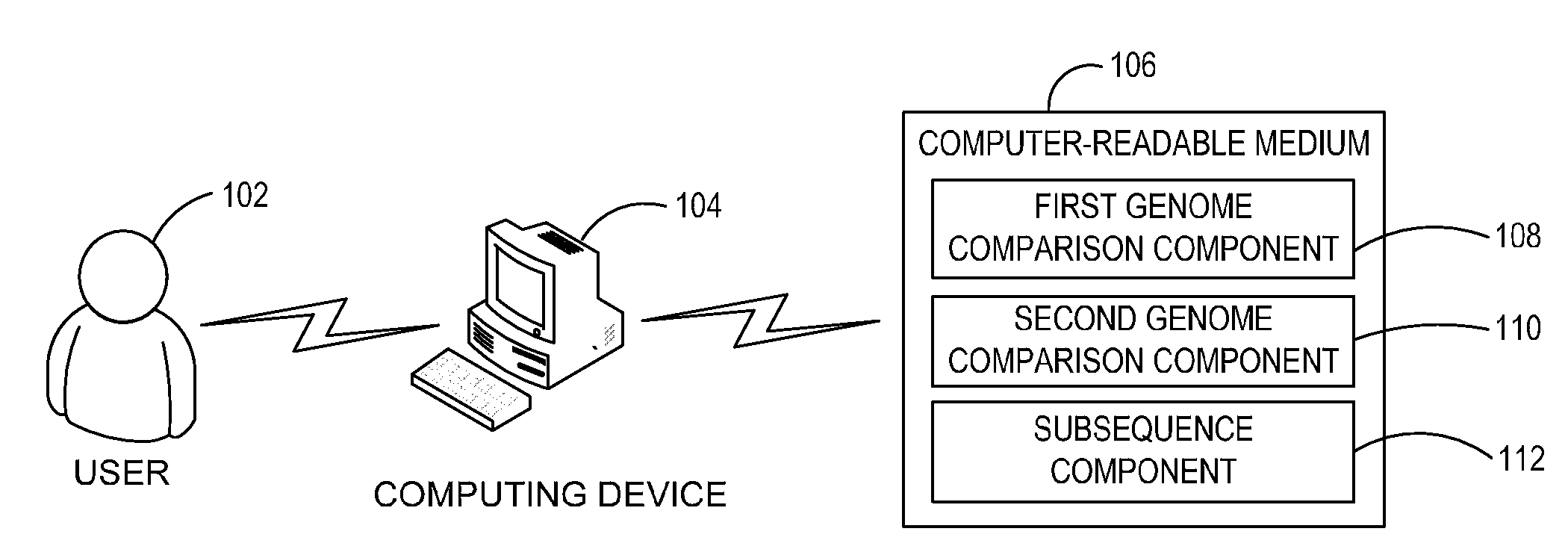
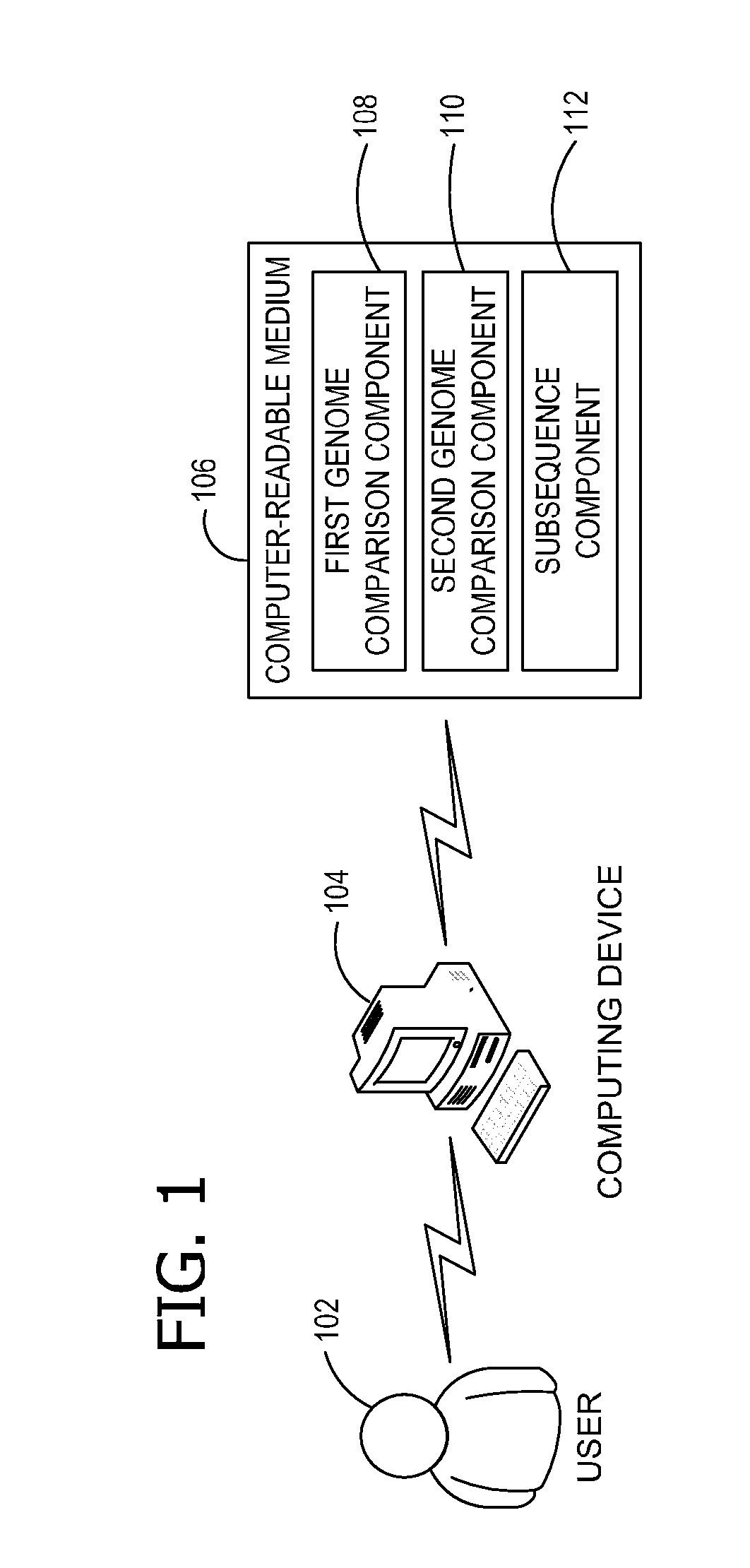
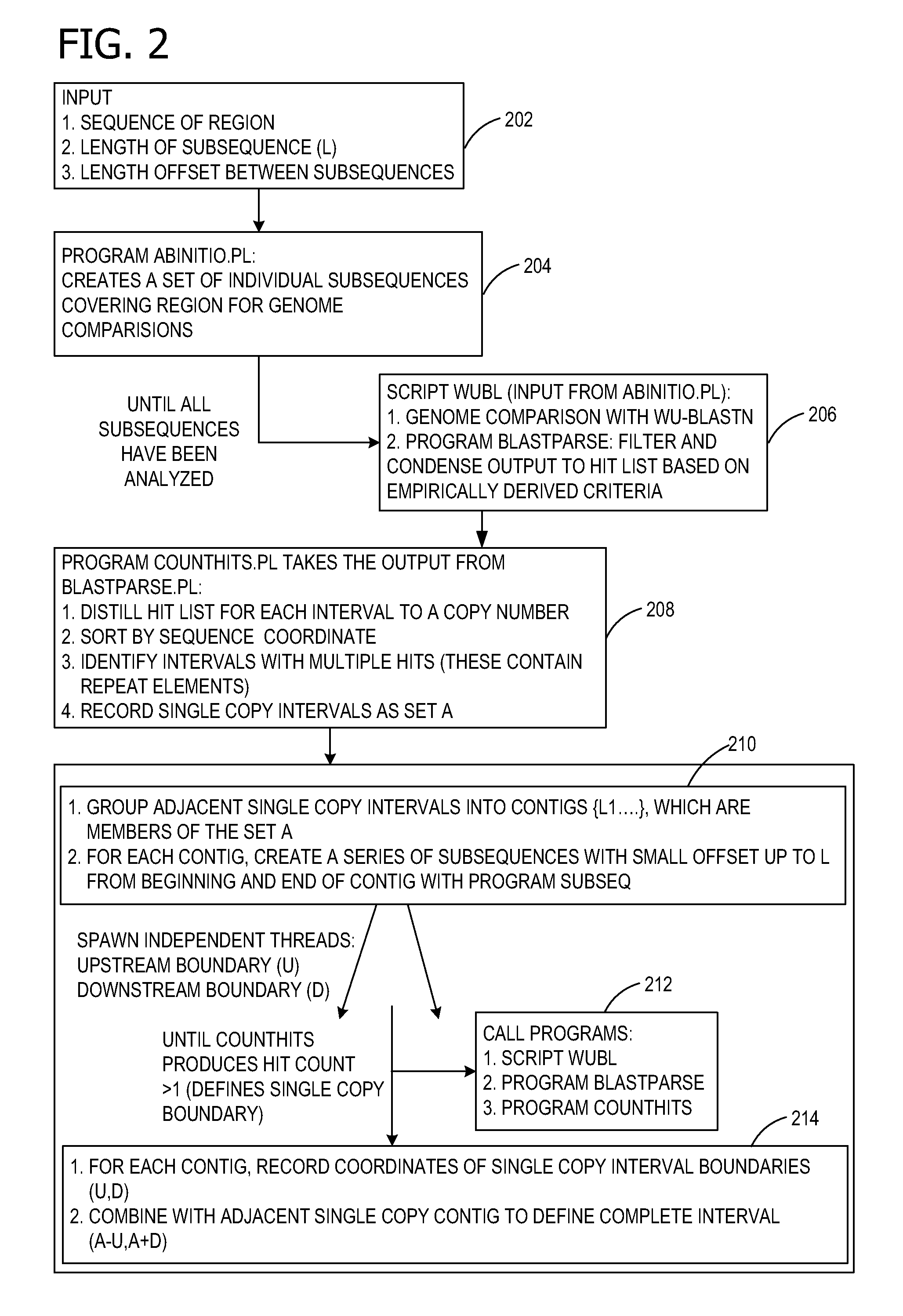
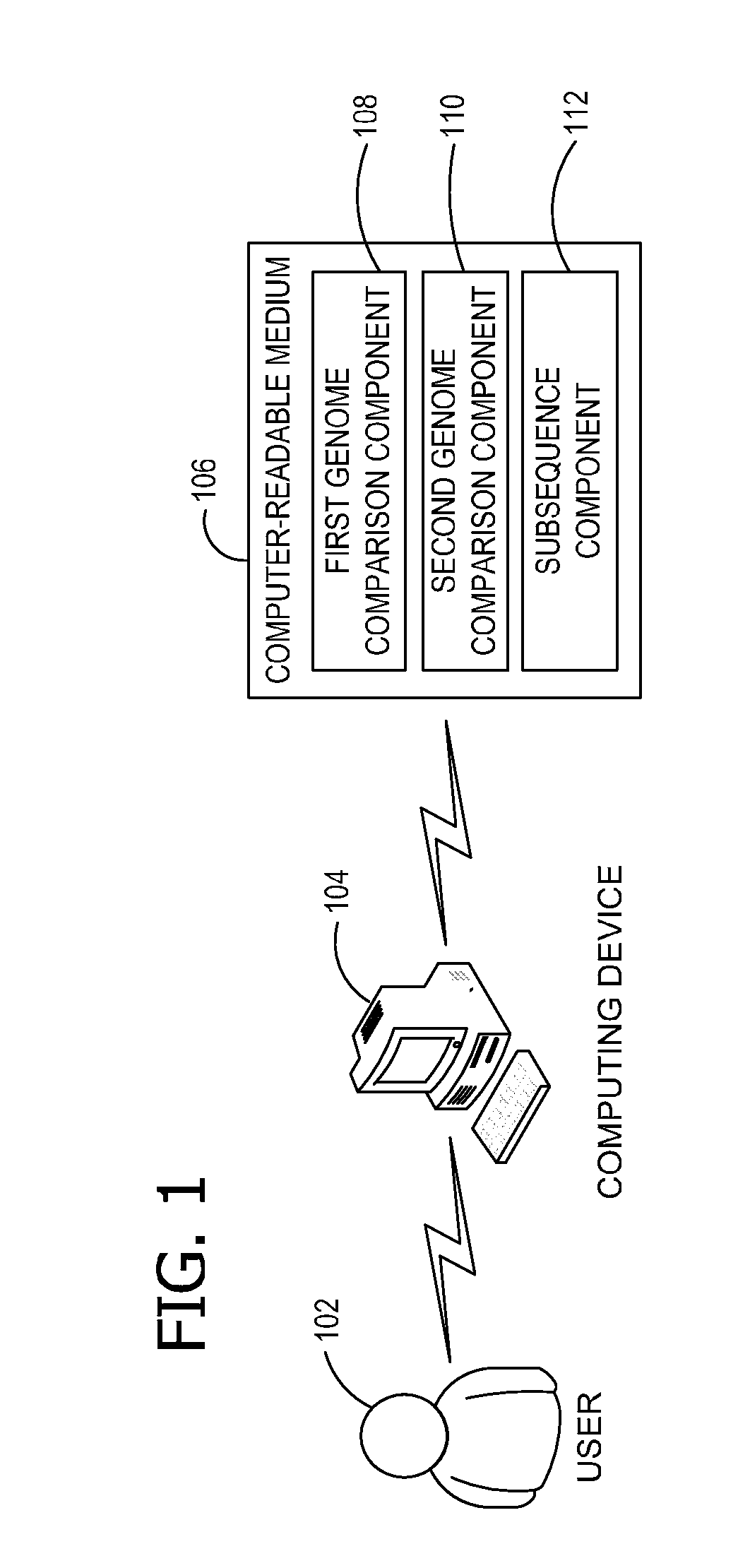
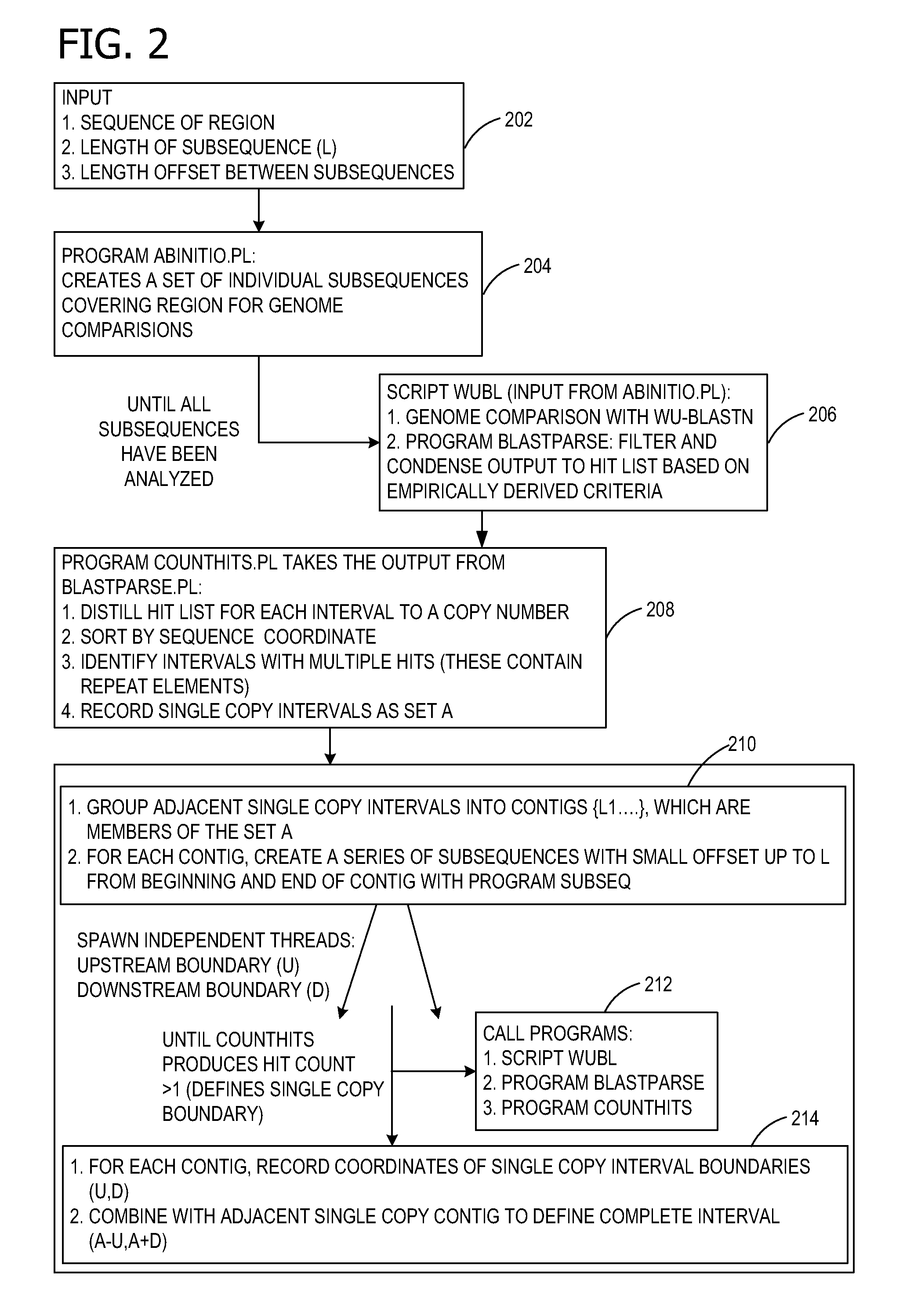
Ab initio generation of single copy genomic probes
Single copy sequences suitable for use as DNA probes can be defined by computational analysis of genomic sequences. The present invention provides an ab initio method for identification of single copy sequences for use as probes which obviates the need to compare genomic sequences with existing catalogs of repetitive sequences. By dividing a target reference sequence into a series of shorter contiguous sequence windows and comparing these sequences with the reference genome sequence, one can identify single copy sequences in a genome. Probes can then be designed and produced from these single copy intervals.
Owner:ROGAN PETER K
Reorganized mannase, genetically-engineered bacteria of recombined mannose and hydrolyzing preparation mannan oligosaccharide method
InactiveCN103146724AEasy to operateReduce manufacturing costFungiMicroorganism based processesEngineered geneticHydrolysis
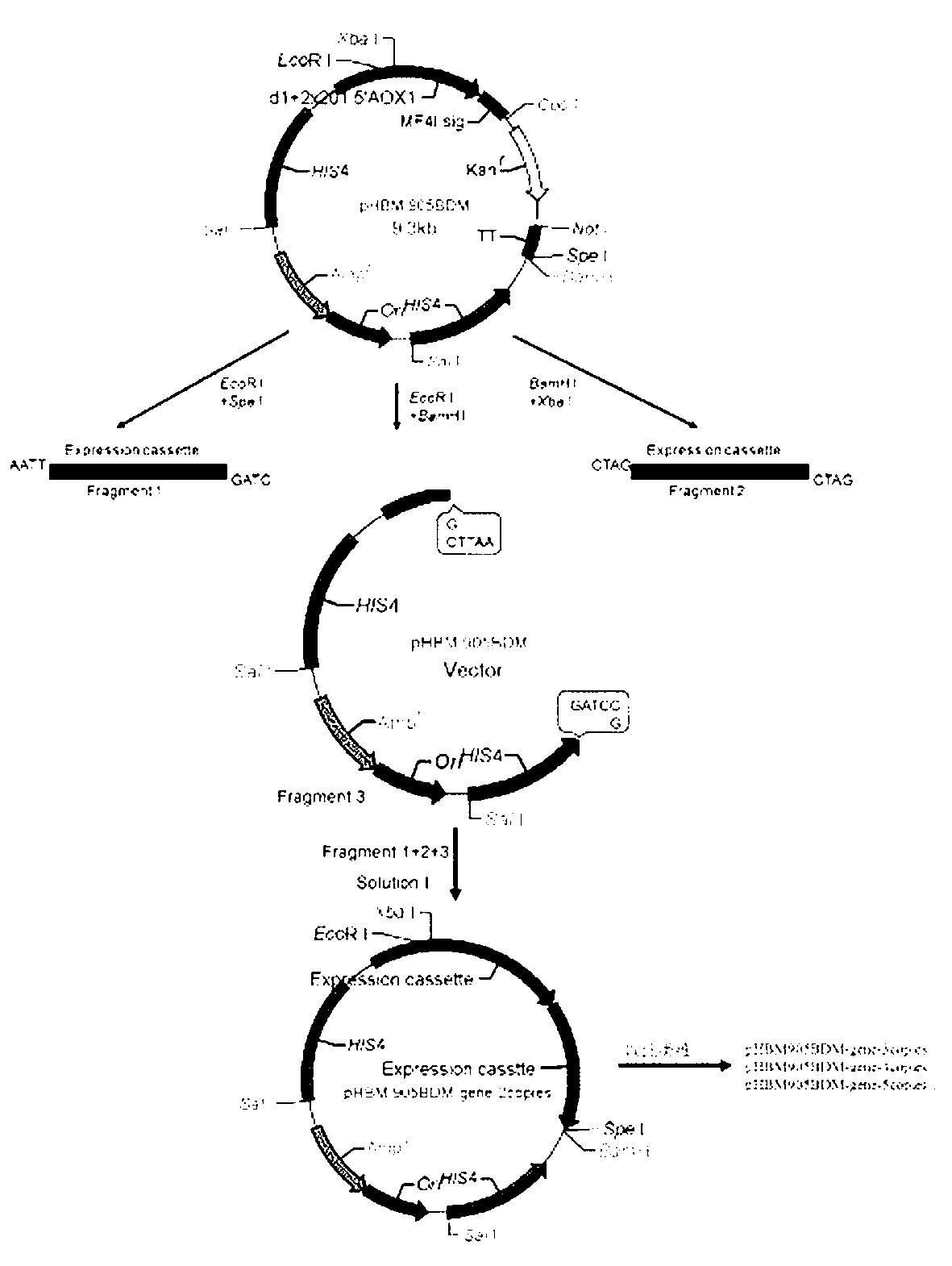
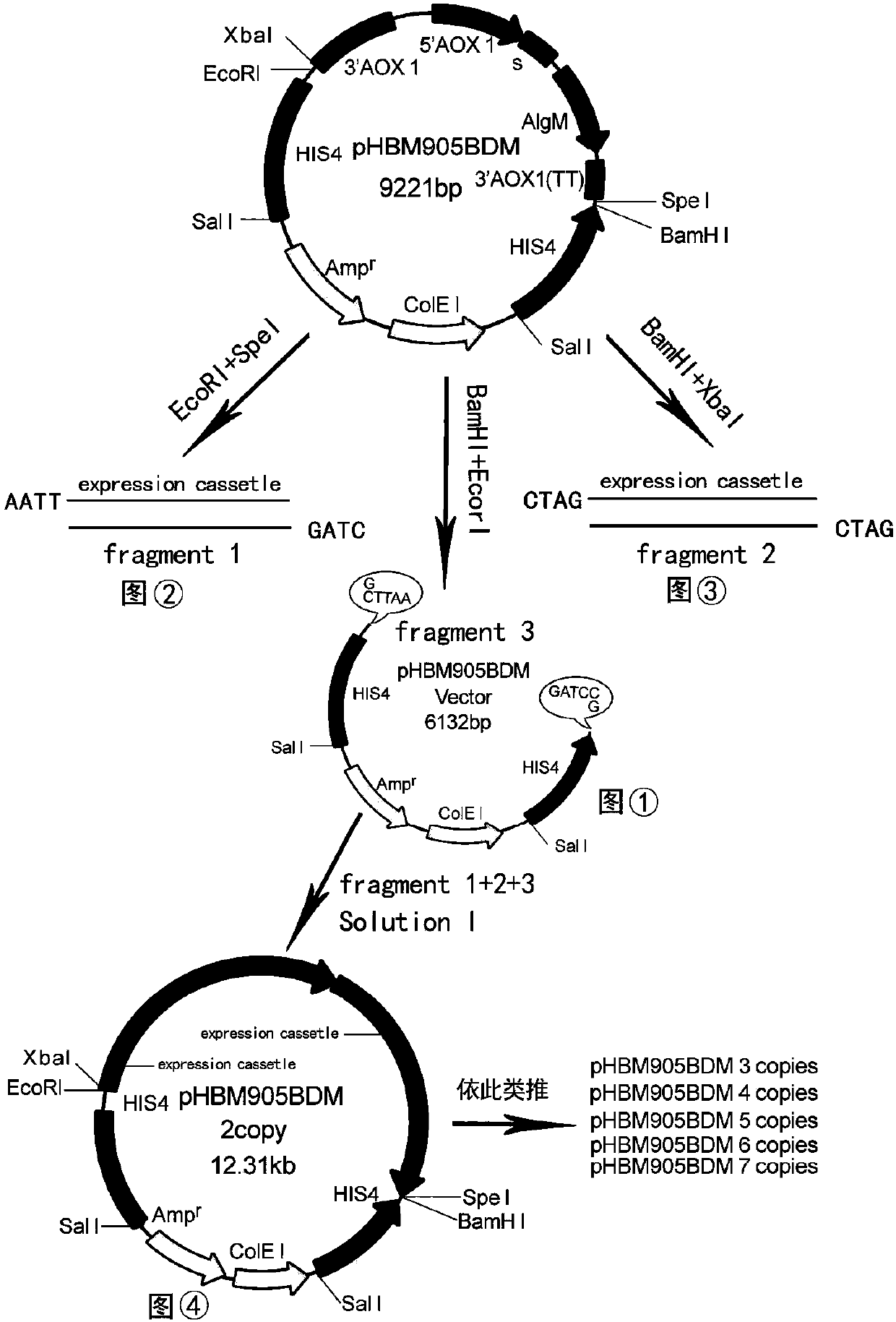
The invention provides reorganized mannase, genetically-engineered bacteria of the recombined mannose and a hydrolyzing preparation mannan oligosaccharide method. The hydrolyzing preparation mannan oligosaccharide method comprises the steps 1), optimizing and reorganizing beta-mannase genes according to pichia pastoris codon preference; 2), constructing expression vector pHBM905BDM-Man containing the beta-mannase genes, converting pitchia pastoris GS115 competence cells, and fostering and checking to obtain single-copy genetically-engineered bacteria; 3) constructing, fostering and checking to obtain multi-copy genetically-engineered bacterium; 4) using different-copy-number genetically-engineered bacteria to efficiently express and prepare reorganized beta-mannase; and 5) using reorganized beta-mannase hydrolysis mannan substrate to produce mannan oligosaccharide. Substrate hydrolysis concentration is 15-40%, temperature is 50 DEG C to 55 DEG C, and time is 0.5 hour to 2 hours. At present, two-copy beta-mannase genetically-engineered bacteria GS115 / MAN78(CCTCCNo:2012554) is high in expression level. The activity of the beta-mannase is 5000U / mL (a decimal number system method), hydrolysis substrate specificity is high, and hydrolysis substrate concentration is obviously higher than that of other methods.
Owner:HUBEI UNIV +1
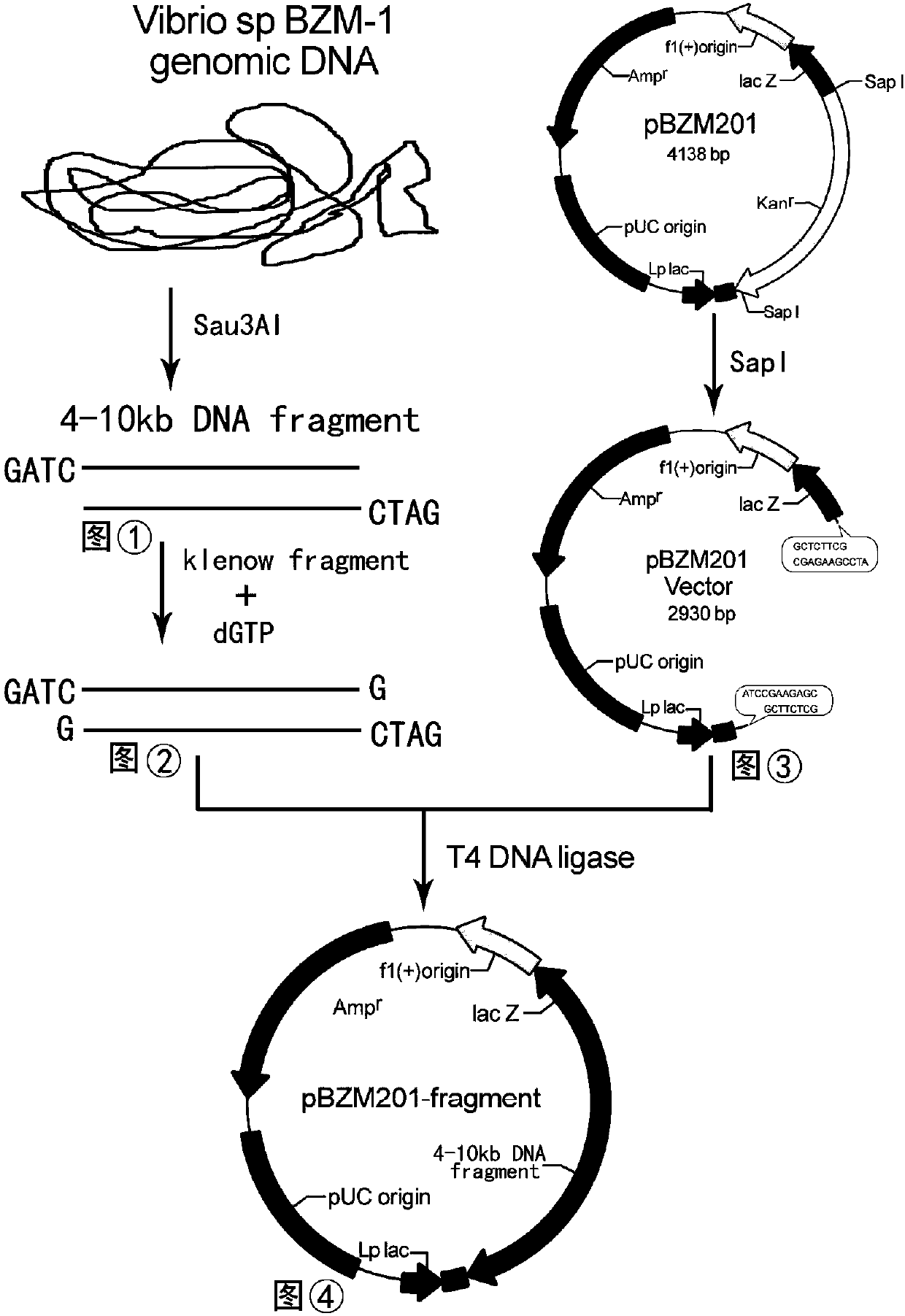
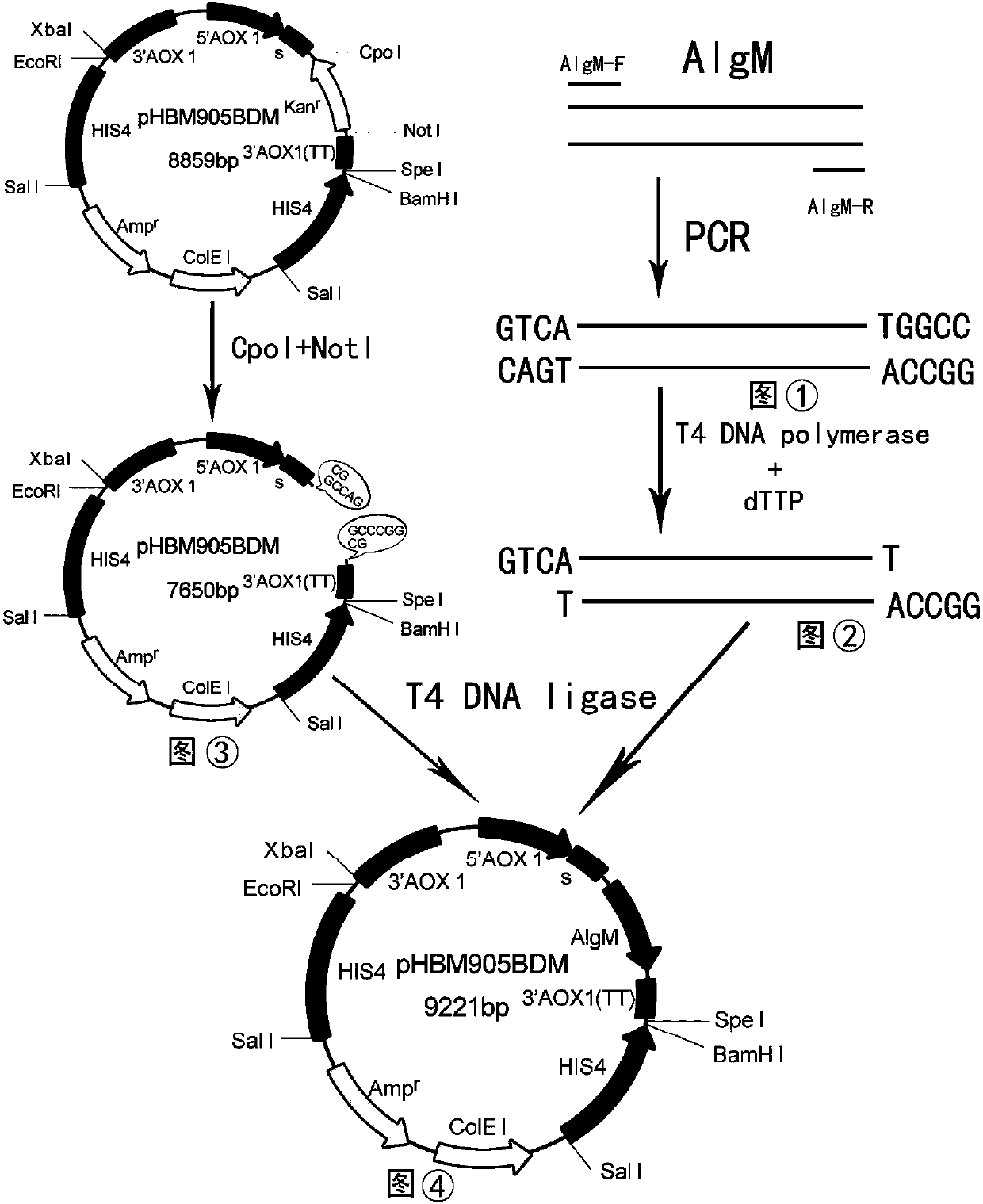
Gene of novel alginate endolyase, engineering bacterium and application
ActiveCN105154458AIncrease enzyme activityHigh purityFungiMicroorganism based processesCompetent cellAlginate lyase
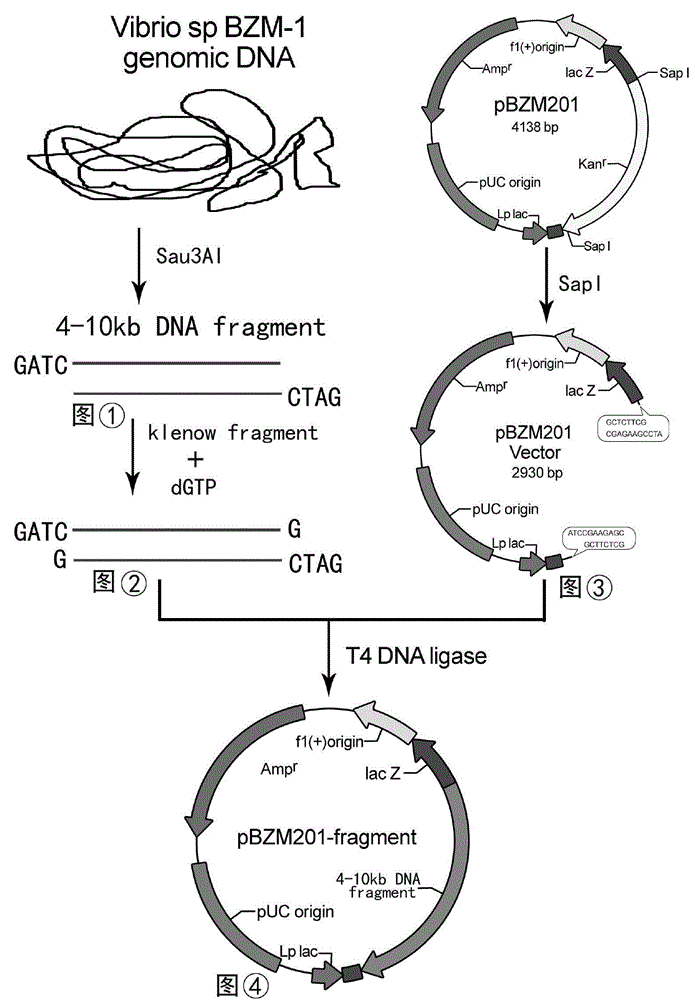
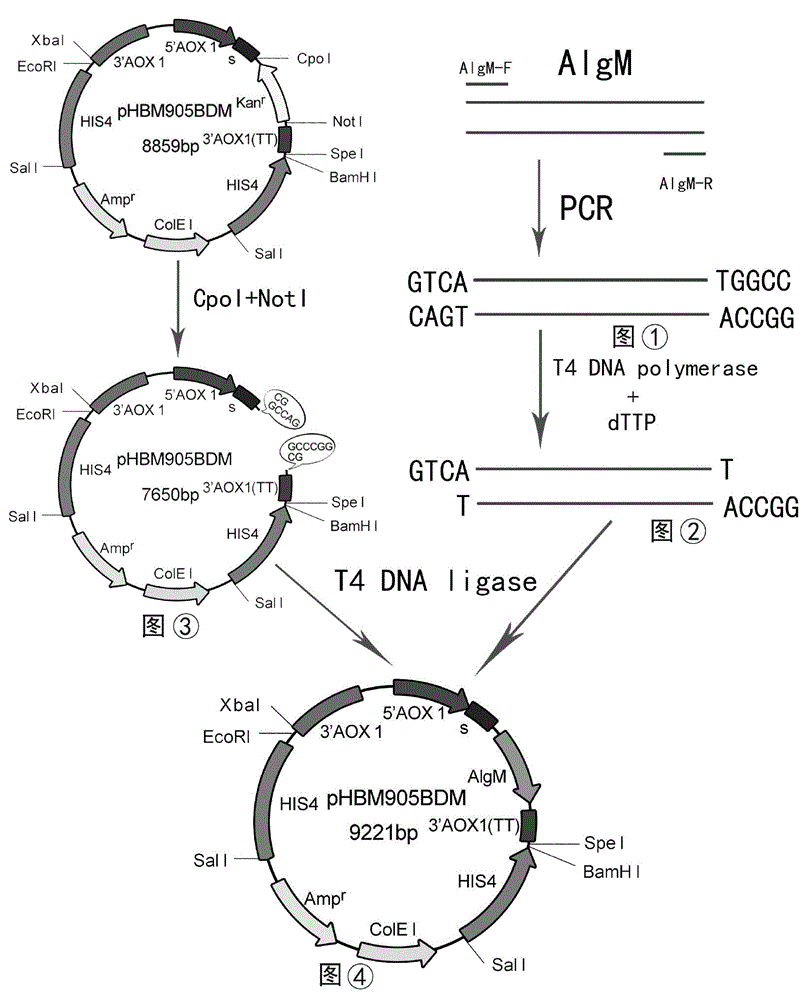
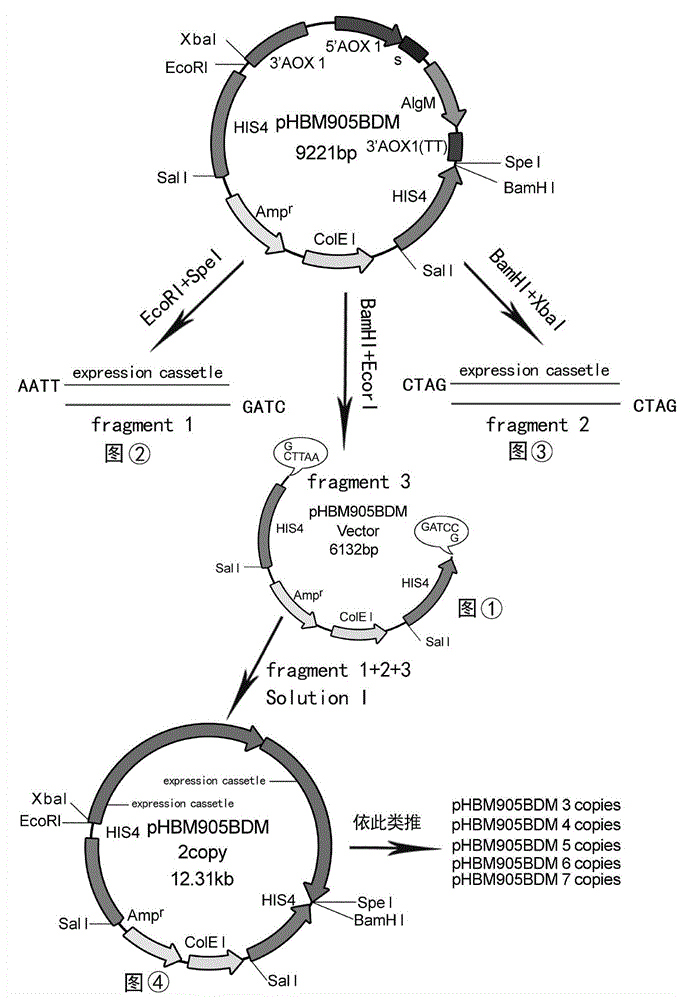
The invention discloses recombinant alginate lyase, an engineering bacterium thereof and a method for preparing alginate oligosaccharide through hydrolysis. The method is characterized by screening an alginate lyase gene AlgM according to a strain Vibrio sp.BZM-1 newly isolated from seaweeds; constructing an expression vector pHBM905BDM-AlgM containing the gene, and converting pichia sp GS115 competent cells to obtain a single copy gene engineering bacterium; and constructing, culturing and verifying the multi-copy gene engineering bacterium to prepare recombinant alginate lyase through high-efficiency expression, and producing alginate oligosaccharide by hydrolyzing sodium alginate with alginate lyase. The recombinant alginate lyase gene engineering bacterium GS115 / AlgM4 (CCTCC No:M2015500) containing four copies has a higher expression level. The activity of recombinant alginate lyase is as high as 31000U / mL.
Owner:BINZHOU MEDICAL COLLEGE
O-type foot-and-mouth disease recombined phage vaccine of pig and construction method thereof
InactiveCN102416174AOvercoming the deficiency of weaker autoimmunogenicityOvercoming the disadvantage of lacking T helper cell epitopesMicroorganism based processesAntiviralsDiseaseT7 phage
The invention provides an O-type foot-and-mouth disease recombined phage vaccine of a pig and a construction method thereof. The vaccine adopts recombined T7 phage as an antigen, and the amino acid sequence such as FMDVVP1(129-169) polypeptide of Mya98 pedigree shown in SEQIDNO:2 is expressed on the surface of the recombined T7 phage; and the VP1 (129-169) polypeptide is obtained by inserting single-copy VP1(129-169) genes into the downstream of the P10B gene of the T7 phage to carry out fusion. The invention has the following technical effects that (1) the recombined phage vaccine prepared by the T7 phage carrier can overcome the shortcomings of weaker self immunogenicity of the FMDVVP1 polypeptide and the defect of lack of T auxiliary cell epitope, effectively improve the immunogenicity and stimulate the organism to generate the higher-level antibody; and (2) since the T7 phage is easy to culture, the purification is convenient, and the invention for producing the gene engineering vaccine has multiple advantages. In the O-type foot-and-mouth disease recombined phage vaccine of the pig, the on-scale production is convenient and the cost is low.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
AB initio generation of single copy genomic probes
ActiveUS8407013B2Sugar derivativesMicrobiological testing/measurementReference genome sequenceAb initio
Single copy sequences suitable for use as DNA probes can be defined by computational analysis of genomic sequences. The present invention provides an ab initio method for identification of single copy sequences for use as probes which obviates the need to compare genomic sequences with existing catalogs of repetitive sequences. By dividing a target reference sequence into a series of shorter contiguous sequence windows and comparing these sequences with the reference genome sequence, one can identify single copy sequences in a genome. Probes can then be designed and produced from these single copy intervals.
Owner:ROGAN PETER K
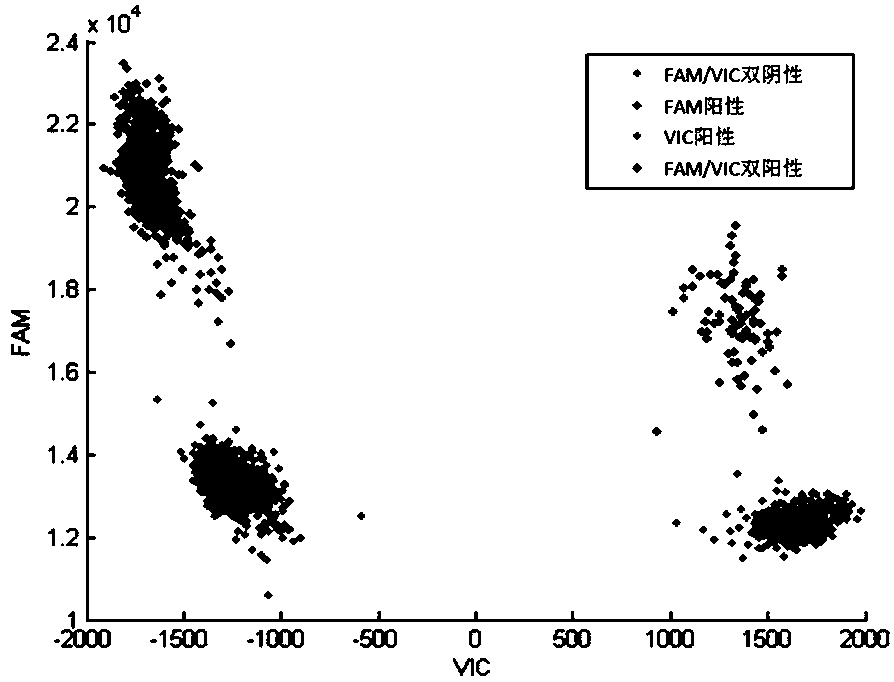
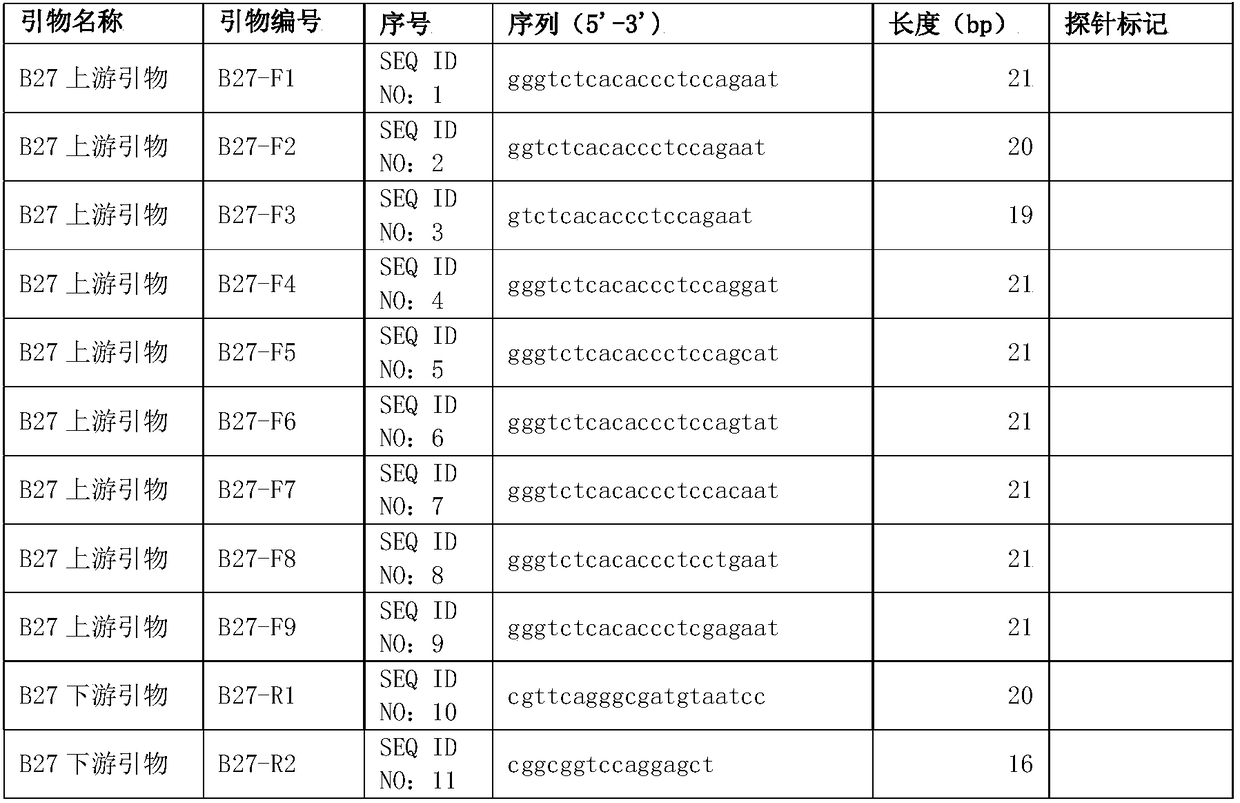
Method of using digital PCR (polymerase chain reaction) to identify single-copy gene mutation genotypes
InactiveCN108384843ARapid identificationReliable identificationMicrobiological testing/measurementHousekeeping geneGenotype
The invention provides a method of using digital PCR (polymerase chain reaction) to identify single-copy gene mutation genotypes, comprising: designing a digital PCR amplification primer and a probe for a sample's single-copy gene mutation site to be detected, designing a digital PCR amplification primer and a probe for the sample's single-copy housekeeping gene, and subjecting the mutation site and the housekeeping gene to dual-channel double digital PCR; determining whether the sample to be detected has the single-copy gene mutation site according to presence or absence of digital PCR amplification of the single-copy gene mutation site; if yes, identifying the genotype of the single-copy gene to be detected by comparing the copy number of the single-copy gene to be detected to the copy number of the single-copy housekeeping gene. The method has the advantages of good accuracy, good convenience, high speed, low cost and the like and is suitable for genetic disease screening and prevention and genetic diagnosis.
Owner:TARGETINGONE TECH (BEIJING) CORP
Method for accurately measuring length of crowd telomere
InactiveCN106755429AExact telomere lengthReduce dosageMicrobiological testing/measurementHuman DNA sequencingLength measurement
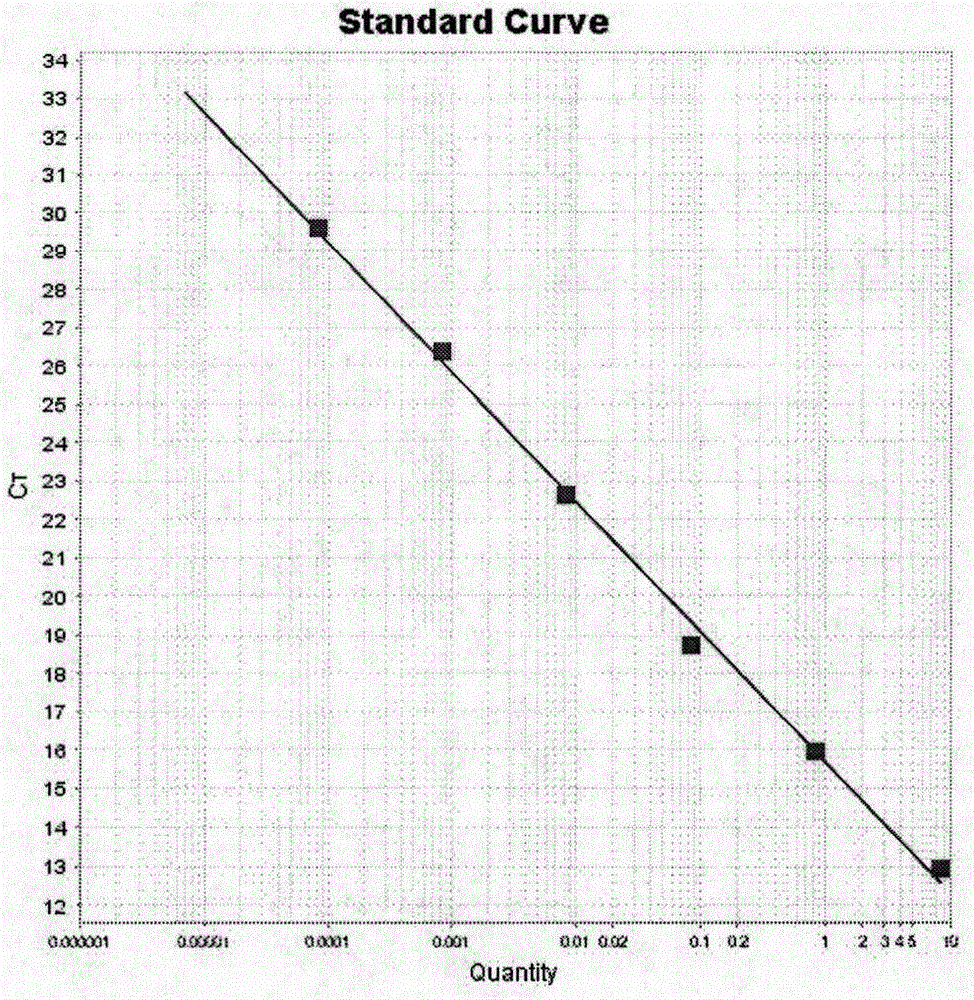
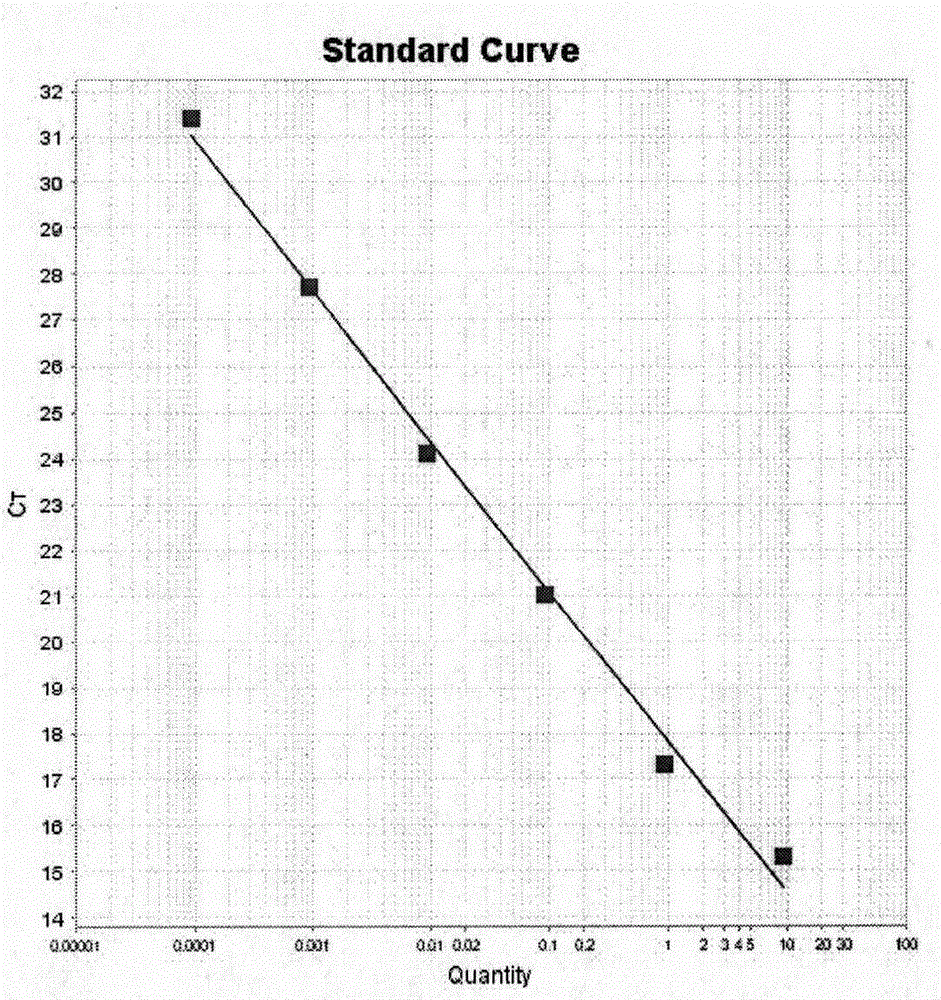
The invention relates to the technical field of molecular organism and specifically relates to a method for accurately measuring length of crowd telomere. A 36B4 gene of a coding acidic ribosome phosphoprotein is selected as a single copy gene. The method comprises the following steps: extracting a human genome DNA; designing a reference substance and a primer and compounding; designing telomere and 36B4 standard curve; adopting a RT-qPCR method for measuring the length of crowd telomere; analyzing data. According to the invention, on the basis of the RT-qPCR method, the telomere standard curve and the single copy gene standard curve are added for more accurately measuring T and S values, so that a more accurate telomere length can be acquired; the DNA dosage is small; only 60ng DNA is required; the measuring speed is high; the measurement has comparability; the telomere length can be accurately measured in batches; and the method is specially suitable for the measurement for the crowd telomere length.
Owner:上海三誉华夏基因科技有限公司
Single copy genomic hybridization probes and method of generating same
InactiveUS20050064449A1Eliminates spurious hybridizationSugar derivativesMicrobiological testing/measurementGenomic SegmentHybridization probe
Nucleic acid (e.g., DNA) hybridization probes are described which comprise a labeled, single copy nucleic acid which hybridizes to a deduced single copy sequence interval in target nucleic acid of known sequence. The probes, which are essentially free of repetitive sequences, can be used in hybridization analyses without adding repetitive sequence-blocking nucleic acids. This allows rapid and accurate detection of chromosomal abnormalities. The probes are preferably designed by first determining the sequence of at least one single copy interval in a target nucleic acid sequence, and developing corresponding hybridization probes which hybridize to at least a part of the deduced single copy sequence. In practice, the sequences of the target and of known genomic repetitive sequence representatives are compared in order to deduce locations of the single copy sequence intervals. The single copy probes can be developed by any variety of methods, such as PCR amplification, restriction or exonuclease digestion of purified genomic fragments, or direct synthesis of DNA sequences. This is followed by labeling of the probes and hybridization to a target sequence.
Owner:KNOLL JOAN +1
Simple and new method for identifying copy numbers of transgenic plants
InactiveCN101445826AEasy to operateReduce the difficulty of testingMicrobiological testing/measurementElectrophoresisAgarose electrophoresis
The invention provides a simple and new method of using a common PCR cycler and a specific single copy gene as interior labels to realize the identification of copy numbers of transgenic plants. The invention is characterized by extracting the total DNA of young leave samples to be detected, adjusting the concentration of the DNA of all the samples to be consistent, designing specific primers of target genes and interior label genes, further determining the best PCR cycle number on the basis of PCR molecular detection, then making parallel PCR and agarose electrophoresis of the target genes and the interior label genes under the same cycle number and finally determining the copy numbers of the target genes according to the ratio of optical density values of the target genes and the interior genes in an electrophoresis image. The invention has the advantage that the invention is beneficial to screening the transgenic plants with stable expression in transgenic plant breeding; the invention has the advantages of simple operation, short time consumption, low cost, safety and accuracy; and the invention combines the PCR molecular detection of the transgenic plants and causes a laboratory for performing the PCR molecular detection of the transgenic plants to realize batch detection without adding experimental equipment and increasing the difficulty.
Owner:新疆农业科学院核技术生物技术研究所
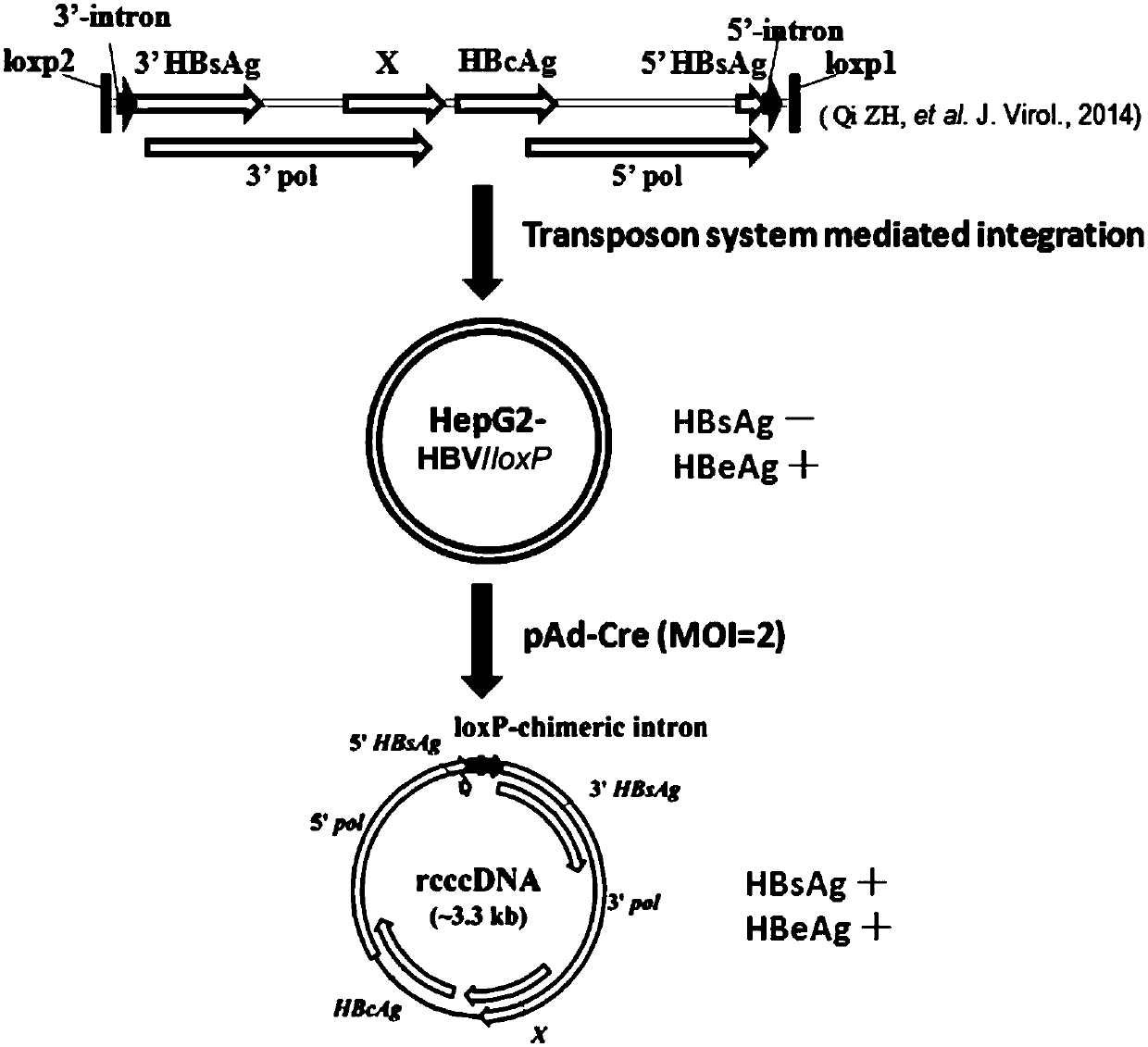
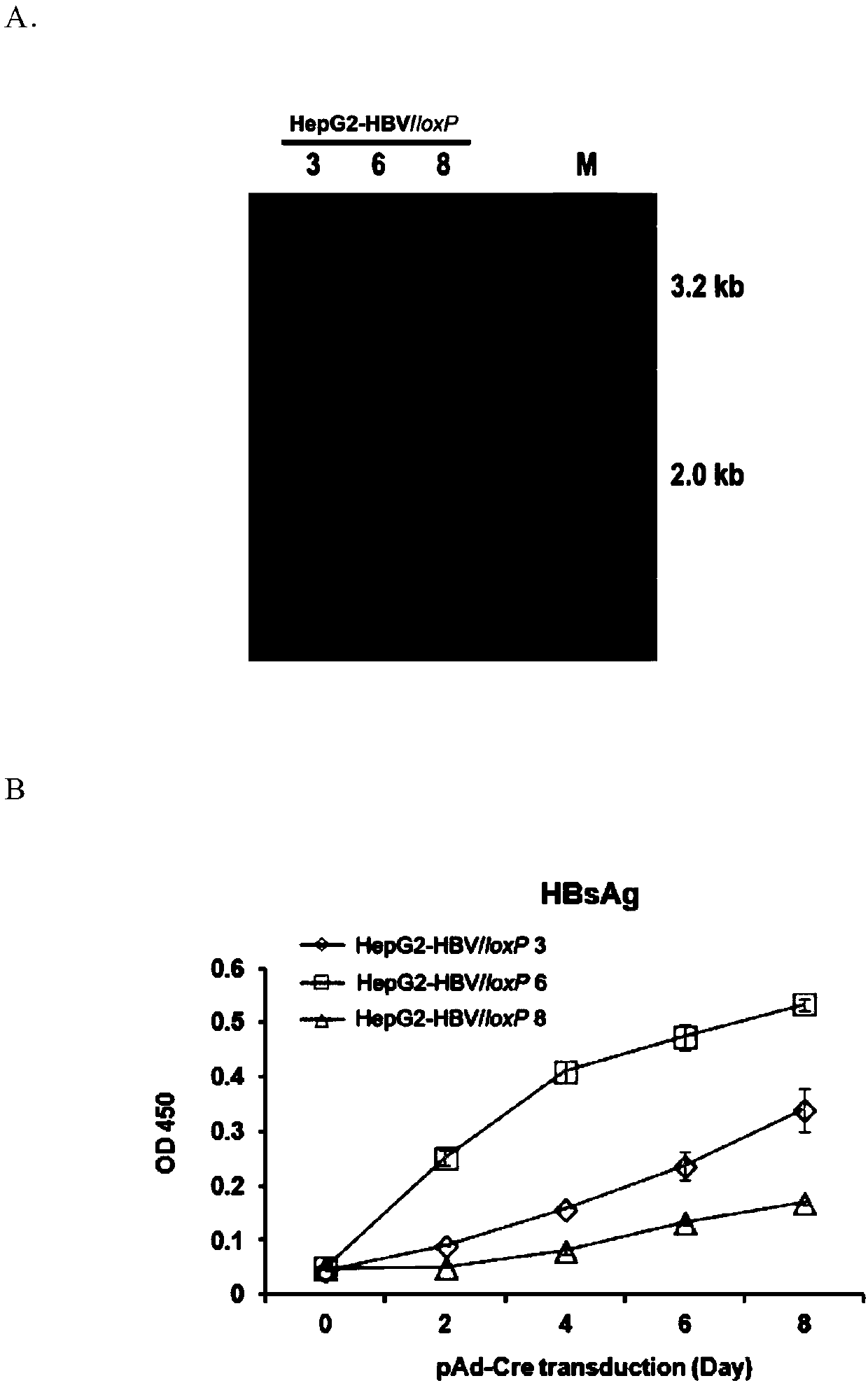
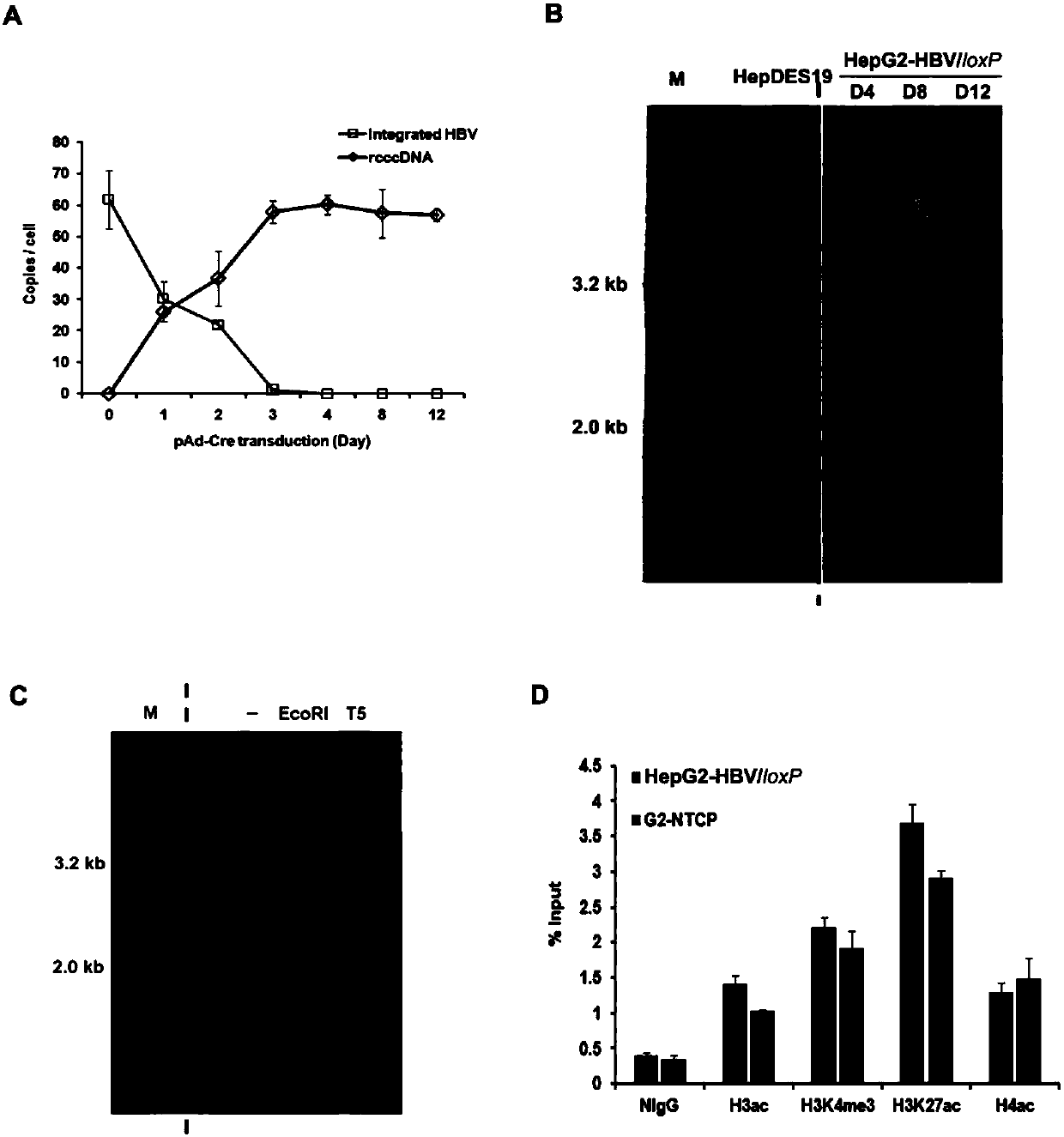
Cell system for simply and efficiently generating hepatitis B virus (HBV) recombinant cccDNA
The invention belongs to the field of microorganism animal cell lines, and relates to a cell system for generating hepatitis B virus (HBV) recombinant cccDNA. Through a transposon system, HBV single-copy genomes with two ends provided with loxP sites are integrated into HepG2 cells, a clone HepG2-HBV / loxP cell line integrated with the different-copy-number HBV genomes is obtained, the cell line does not express HBsAg, after cyclization recombination enzyme (Cre) is introduced through adenovirus transduction, the rcccDNA is generated and the HBsAg is expressed, and expression of the HBsAg is closely relevant to the generation and level of the rcccDNA; the rcccDNA is rapidly and stably generated, the structure feature of the rcccDNA is similar to that of real HBV cccDNA, and transcription, copy and protein expression of viruses can be supported; the rcccDNA can be quantitatively detected through quantitative PCR by designing a specific primer, and Southern Blot detection can be carried out by utilizing a digoxin marking system. The cell system can be used for establishing a platform for HBV cccDNA relevant biological research and screening and evaluation of anti-HBV medicines.
Owner:FUDAN UNIV +1
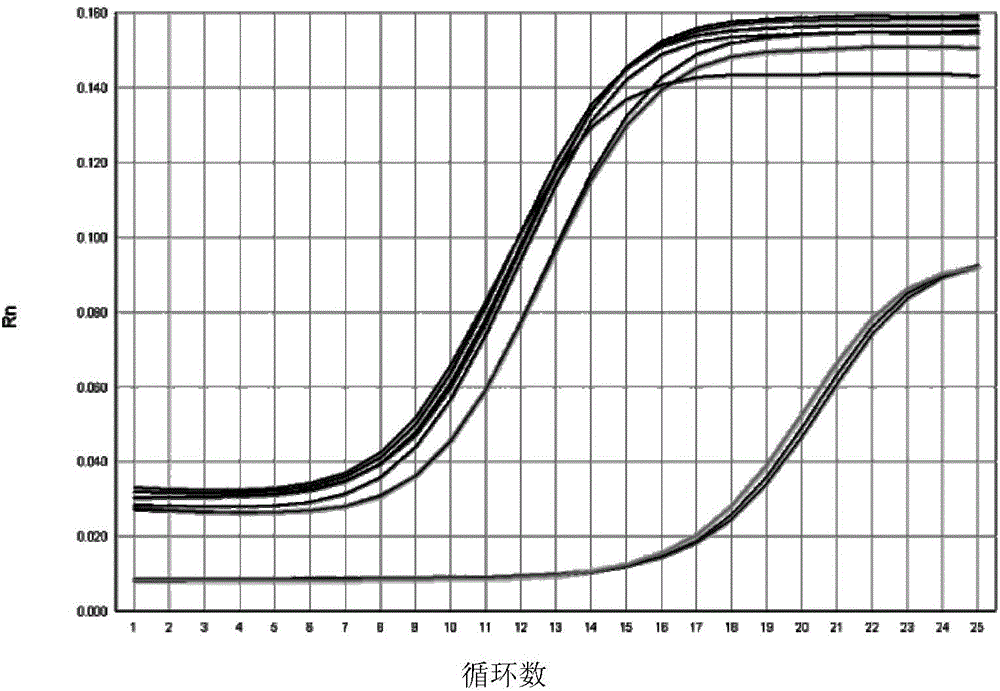
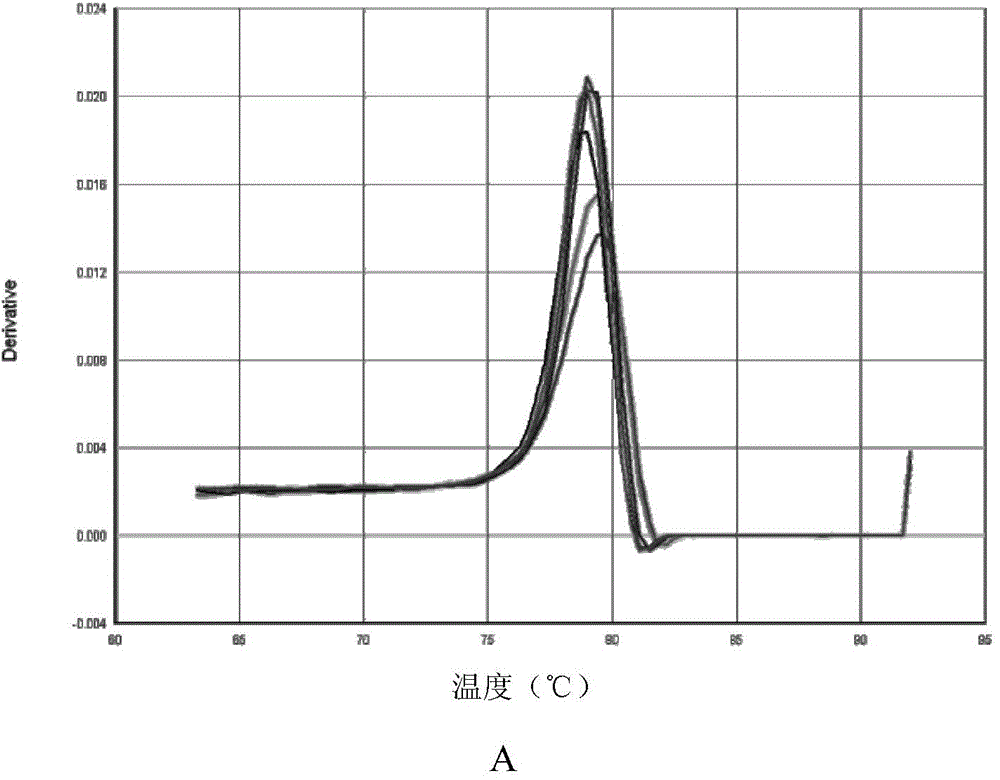
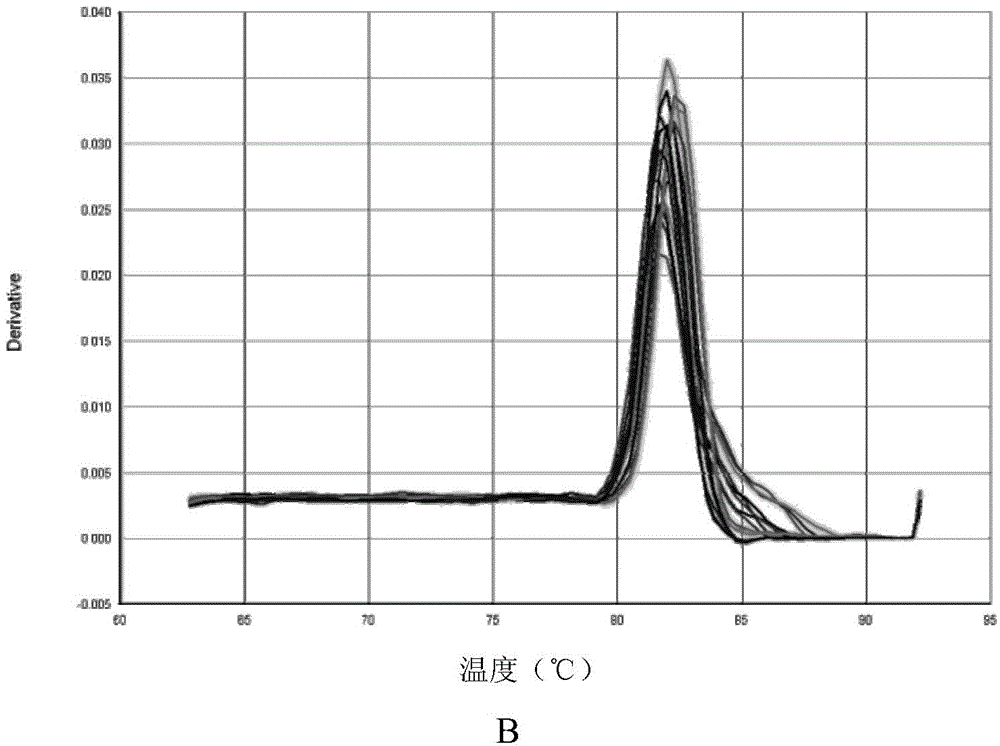
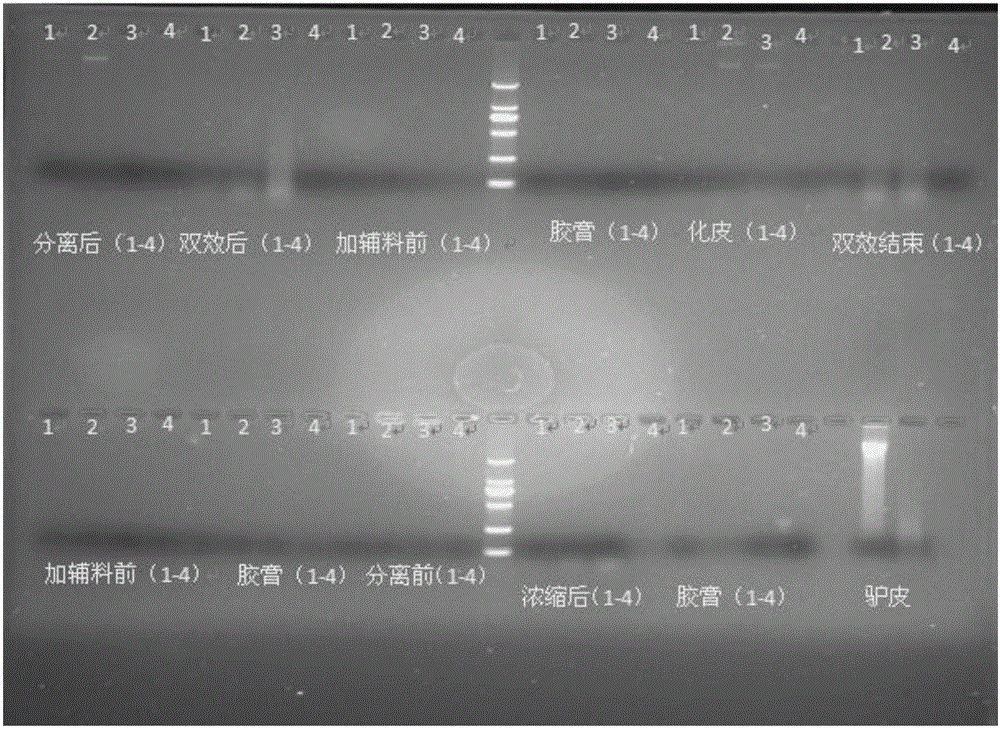
Quantitative detection method, composition and kit of donkey/horse-derived components in donkey-hide gelatin colloidal liquid semifinished product or finished product
InactiveCN105821129AImprove detection success rateImprove integrityMicrobiological testing/measurementDNA/RNA fragmentationUnit massWork in process
The invention relates to a quantitative detection method, composition and kit of donkey / horse-derived components in a donkey-hide gelatin colloidal liquid semifinished product or finished product, belonging to the technical field of molecular biology. The method comprises the following steps: designing donkey / horse universal primers and donkey / horse specific probes according to the donkey / horse house-keeping gene single-copy genes, extracting DNA (deoxyribonucleic acid) of a sample to be detected, carrying out TaqMan qPCR (quantitative polymerase chain reaction) on the donkey-hide gelatin colloidal liquid semifinished product or finished product to perform quantitative detection on donkey / horse-derived components, calculating the number of copies of the donkey / horse-derived nuclear genes in the unit mass of the donkey-hide gelatin colloidal liquid according to the standard curve and the Ct value of the sample to be detected, and calculating the proportion of the donkey / horse-derived components in the donkey-hide gelatin, thereby quantifying the donkey / horse-derived components in the donkey-hide gelatin sample. The method, composition and kit have the advantages of high specificity and high sensitivity, implement qualitative detection on the donkey / horse-derived components in the donkey-hide gelatin, and can also implement quantitative analysis, thereby providing technological popularization and application for quality control of donkey-hide gelatin enterprises.
Owner:BIOTECH RES CENT SHANDONG ACADEMY OF AGRI SCI
Pork component identifying molecular marker and application thereof
ActiveCN110373473AQuantitatively detectableAvoid pollutionMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyPork meat
The invention discloses a pork component identifying molecular marker and application thereof. According to a SYBR Green I dye based real-time fluorescence quantitative PCR technology, a high-specificity and high-sensitivity pork component detection method is created by taking a pig single-copy gene in a cell nucleus as the molecular marker. By adoption of the method, pigs can be accurately distinguished from other 13 animals and 5 plants, and by practical detection of 6 meat products on the market, whether porcine-derived components are contained or not can be accurately judged. The porcine specific real-time fluorescence quantitative PCR detection method provides technical references for qualitative and quantitative detection of porcine-derived components in mixed meat products.
Owner:SOUTH CENTRAL UNIVERSITY FOR NATIONALITIES
Quick and batch test method for copy number of multi-copy genes of genomes
InactiveCN103103280AImprove design requirementsImprove accuracyMicrobiological testing/measurementFluorescence/phosphorescenceBiologyNucleic acid sequence
The invention discloses a quick and batch test method for the copy number of multi-copy genes of genomes by virtue of a PCR (polymerase chain reaction) technology. The method comprises the following steps of: respectively carrying out PCR amplification and product purification on some multi-copy genes and some known single-copy gene on a same tissue DNA (deoxyribonucleic acid) sample to test the mass concentration of each gene PCR purified product, converting the mass concentration into molar concentration according to the nucleotide sequence of the product, and preparing the standard curves of the genes by same RT-PCR (reverse transcription-polymerase chain reaction) by taking the PCR product and the tissue DNA sample which are subjected to gradient dilution as a template, thus obtaining the concentration of each gene in the DNA sample. Under the same extraction efficiency, the copy number of the multi-copy genes is the number obtained by dividing the quantity of the multi-copy genes under the same volume by the quantity of the single-copy gene, so that the copy number of seven different multi-copy genes in same biont can be tested by once 96-pore RT-PCR reaction, and the copy number of eleven different multi-copy genes can be tested by once 384-pore RT-PCR reaction. Therefore, the quick and batch test efficiency can be realized.
Owner:SICHUAN AGRI UNIV
Single copy genomic hybridization probes and method of generating same
InactiveUS20080085509A1Eliminates spurious hybridizationSugar derivativesMaterial analysis by observing effect on chemical indicatorGenomic SegmentHybridization probe
Nucleic acid (e.g., DNA) hybridization probes are described which comprise a labeled, single copy nucleic acid which hybridizes to a deduced single copy sequence interval in target nucleic acid of known sequence. The probes, which are essentially free of repetitive sequences, can be used in hybridization analyses without adding repetitive sequence-blocking nucleic acids. This allows rapid and accurate detection of chromosomal abnormalities. The probes are preferably designed by first determining the sequence of at least one single copy interval in a target nucleic acid sequence, and developing corresponding hybridization probes which hybridize to at least a part of the deduced single copy sequence. In practice, the sequences of the target and of known genomic repetitive sequence representatives are compared in order to deduce locations of the single copy sequence intervals. The single copy probes can be developed by any variety of methods, such as PCR amplification, restriction or exonuclease digestion of purified genomic fragments, or direct synthesis of DNA sequences. This is followed by labeling of the probes and hybridization to a target sequence.
Owner:KNOLL JOAN H M +1
Method for identifying age of ginseng by utilizing length of ginseng telomere
ActiveCN104357560AHigh degree of fitImprove throughputMicrobiological testing/measurementMathematical modelGenomic DNA
The invention discloses a method for identifying the age of ginseng by utilizing the length of a ginseng telomere. The method comprises the following steps: (A) establishing a mathematical model: (a1) selecting a plurality of samples from the positions 1-2 cm below the rhizomes of the ginseng of known age and extracting genomic DNA from each sample; (a2) taking each genomic DNA as a template, conducting qPCR on a primer pair distinguished from the ginseng telomere to obtain a Ct value and marking the Ct value as T, and conducting qPCR on a primer pair distinguished from a single copy gene of the ginseng to obtain another Ct value and marking the Ct value as S; (a3) calculating the T / S ratio for each ginseng; (a4) taking the ages of the ginseng as the abscissa values and taking all the calculated T / S ratios as the ordinate values to draw a standard linear curve and obtain a curve equation; (B) adopting the mathematical model to identify the age of ginseng to be measured: obtaining the T / S ratio for the ginseng to be measured according to the operations from step (a1) to step (a3), and substituting the T / S ratio into the mathematical model to obtain the age of the ginseng to be measured. The method has the advantages that the identified age of ginseng is highly identical with the actual age of the ginseng, the cost is low, and the flux is high. Compared with the traditional Southern blot technology adopted for measuring the length of a ginseng telomere, the method provided by the invention has less demand for DNA and a shorter test period.
Owner:INST OF CHINESE MATERIA MEDICA CHINA ACAD OF CHINESE MEDICAL SCI
Quantitative detection method for donkey-derived and swine-derived components in donkey-hide gelatin liquid semi-finished product or finished product, composition and kit
InactiveCN105803086AOvercome the difficulty of extracting incomplete puzzlesRealize quantitative detectionMicrobiological testing/measurementDNA/RNA fragmentationQuality controlGelatin
The invention relates to a quantitative detection method for donkey-derived and swine-derived components in a donkey-hide gelatin liquid semi-finished product or finished product, a composition and a kit, which belongs to the technical field of molecular biology. The method comprises: according to a donkey and swine house-keeping gene single-copy gene, designing a universal donkey and swine primer as well as a donkey and swine specificity probe, extracting a sample DNA to be tested, quantitatively detecting the donkey-derived or swine-derived components in the donkey-hide gelatin liquid semi-finished product or finished product by adopting a TaqMan qPCR method, calculating a copy number of donkey-derived and swine-derived karyogene in the unit mass of the donkey-hide gelatin liquid according to a standard curve and a Ct value of a sample to be tested, calculating a ratio of the donkey-derived components and a ratio of the swine-derived components in donkey-hide gelatin, and quantifying the donkey-derived or swine-derived components in the gelatin sample. The method, the composition and the kit are good in detection specificity, high in sensitivity, not only capable of qualitatively detecting the donkey-derived and swine-derived components in the donkey-hide gelatin, but also capable of realizing the quantitative analysis, and capable of providing technical popularization and application for the quality control of donkey-hide gelatin enterprises.
Owner:BIOTECH RES CENT SHANDONG ACADEMY OF AGRI SCI
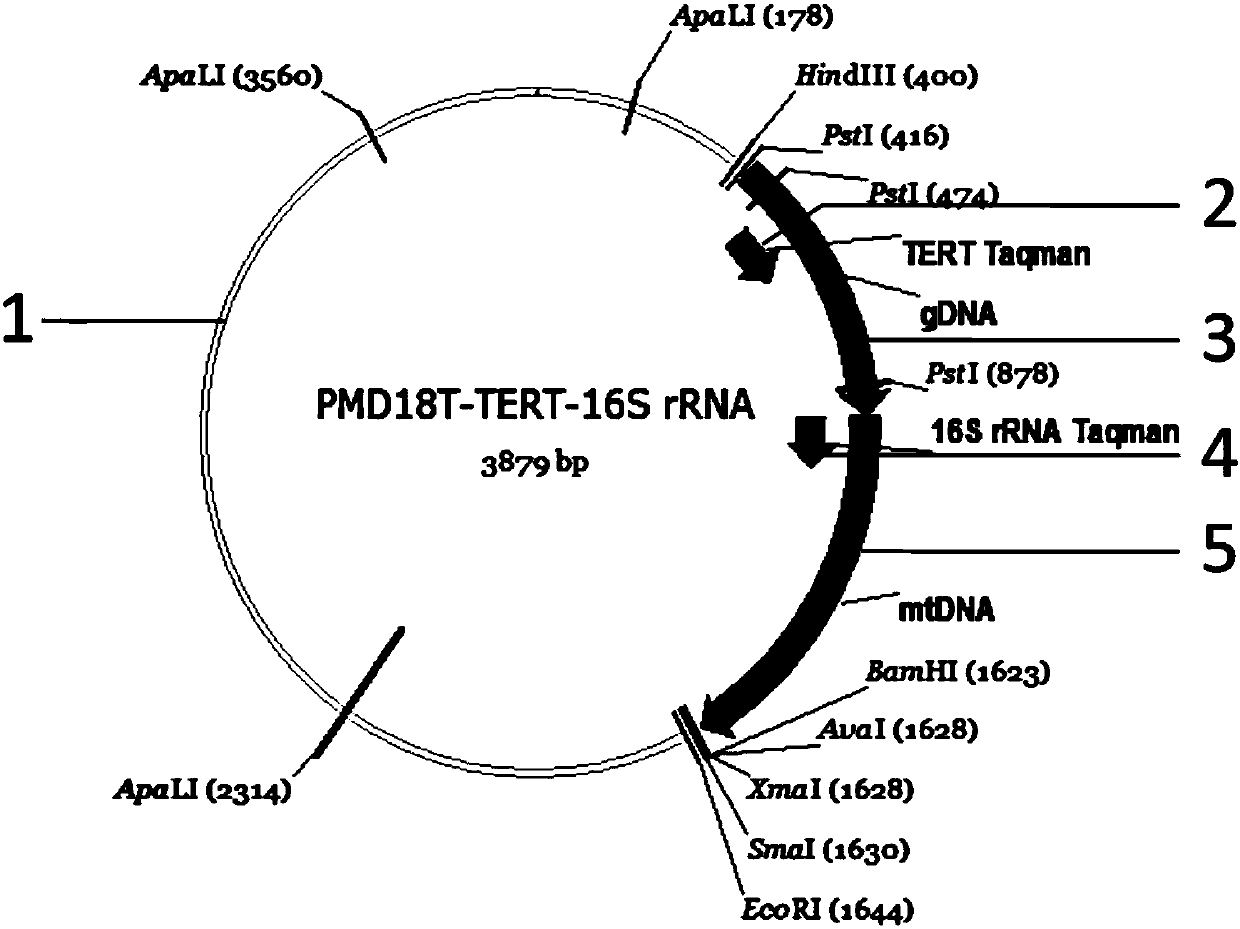
Method for quantitatively detecting nuclear genome copy number and human mitochondrial genome copy number by fluorescence
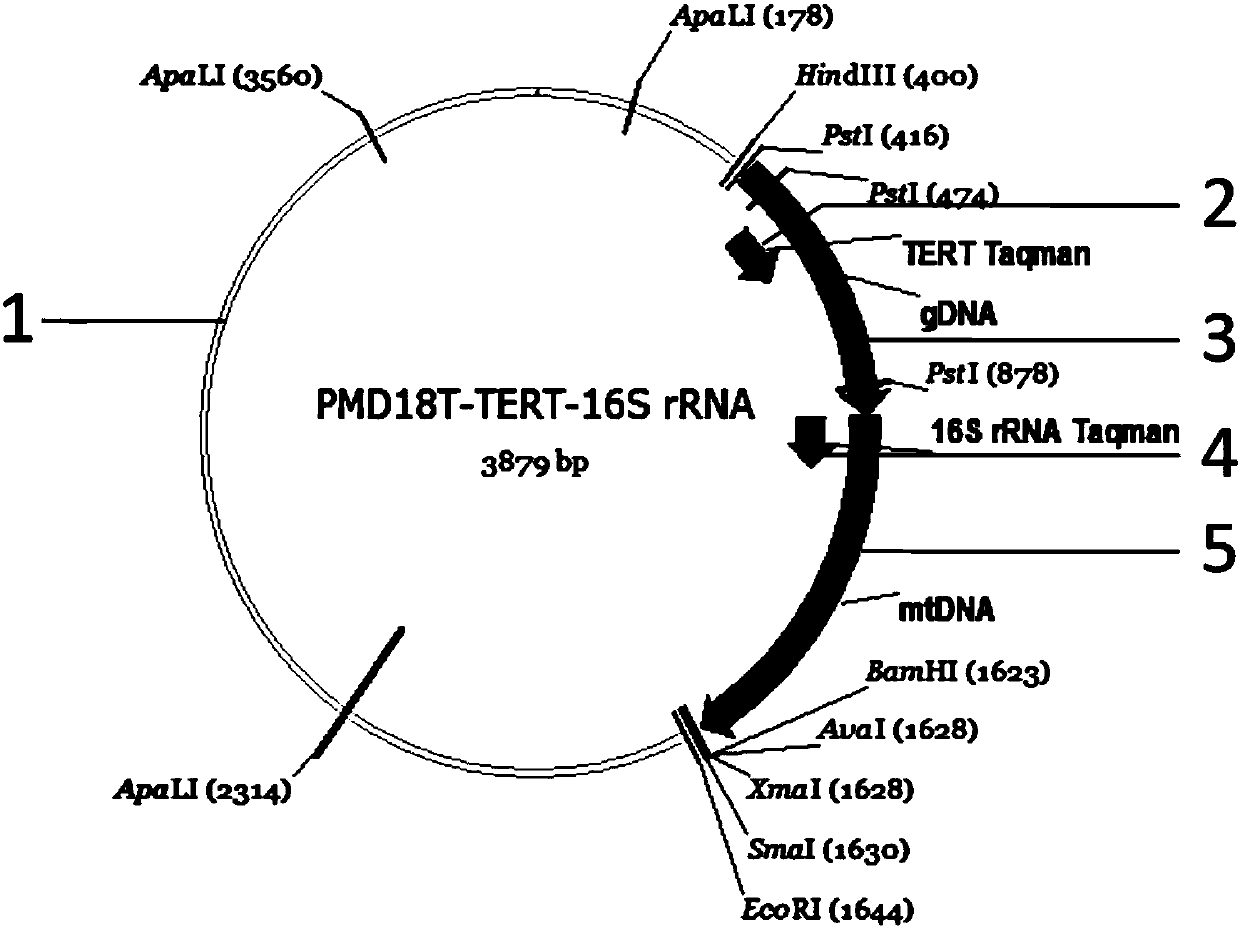
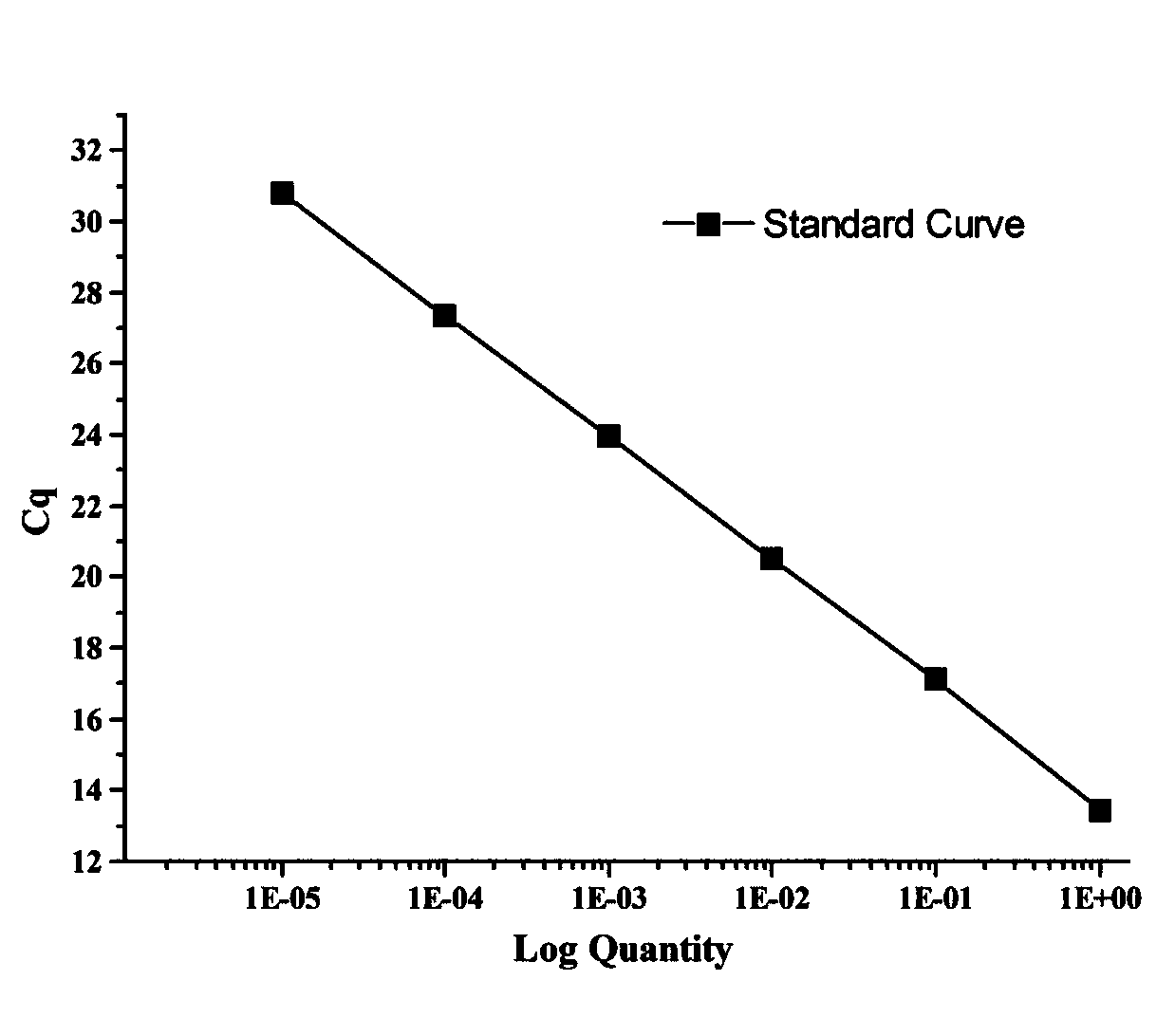
InactiveCN109913535AQuick checkStrong specificityMicrobiological testing/measurementFluorescenceQuantitative PCR instrument
The invention discloses a method for quantitatively detecting nuclear genome copy number and human mitochondrial genome copy number by fluorescence. The method includes respectively selecting a singlecopy gene in human nuclear genome DNA and mitochondrial genome, designing primers and probes, transferring sequence fragments into PMD-18T vectors, extracting plasmids after conversion to obtain thecopy number and taking the copy number as a reference standard; and in actual detection, taking the Cq value of the reference standard to make a standard curve to compare and obtain the copy number ofnuclear genome DNA and mitochondrial genome DNA in a sample to be detected. The method provides a method for quantitatively analyzing the copy number of the mitochondrial genome in different human tissue cells, has high specificity and simple operation, and can complete rapid detection of the copy number of the mitochondrial genome only by a fluorescence quantitative PCR instrument.
Owner:SHANGHAI CRIMINAL SCI TECH RES INST
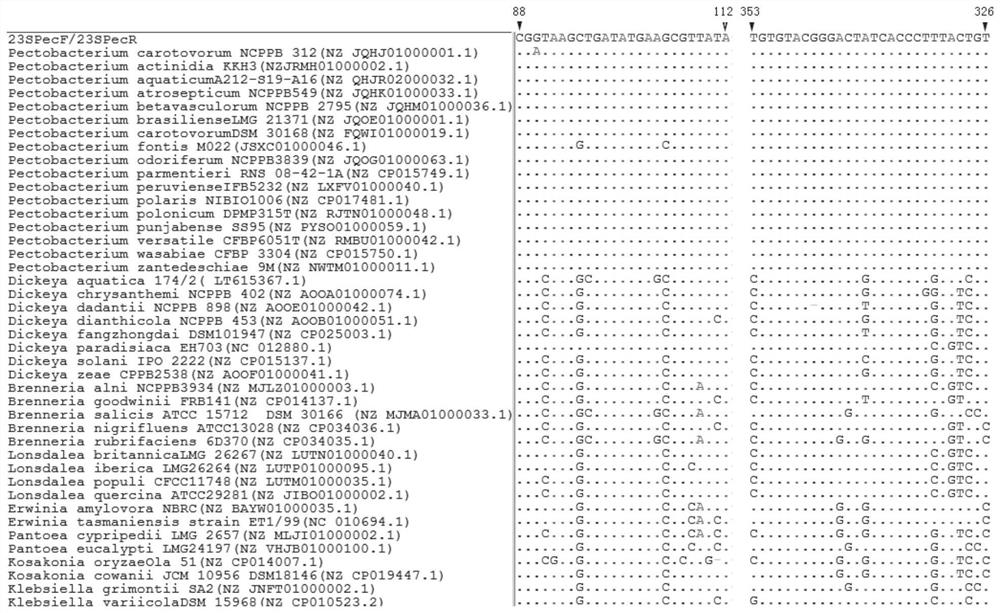
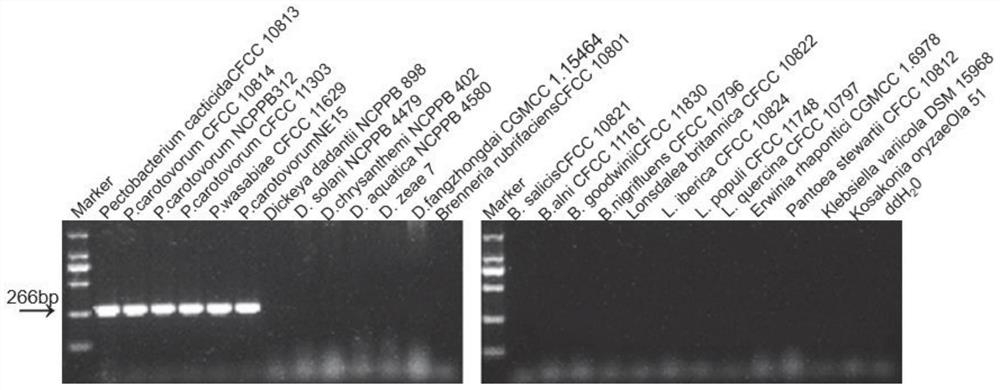
Primer pair, kit and method for detecting and identifying Pectobacterium bacteria
ActiveCN112626241AAccurate identificationMicrobiological testing/measurementMicroorganism based processesForward primerMicrobiology
The invention discloses a primer pair for detecting and identifying Pectobacterium bacteria and a using method thereof. The primer comprises a forward primer 23SPecF and a reverse primer 23SPecR. The method for detecting and identifying the Pectobacterium bacteria comprises the following steps of: performing Polymerase Chain Reaction (PCR) on the 23SPecF and the 23SPecR by using the primer pair, wherein a 266bp nucleic acid fragment can be only amplified from a sample containing the genome DNA of the Pectobacterium bacteria. Due to that the Pectobacterium bacteria have seven copied 23SrRNA genes, the detection sensitivity of the primer pair for performing the PCR on the 23SPecF and the 23SPecR is seven times that of a single-copy gene primer, and the Pectobacterium bacteria can be detected more sensitively.
Owner:ZHEJIANG UNIV
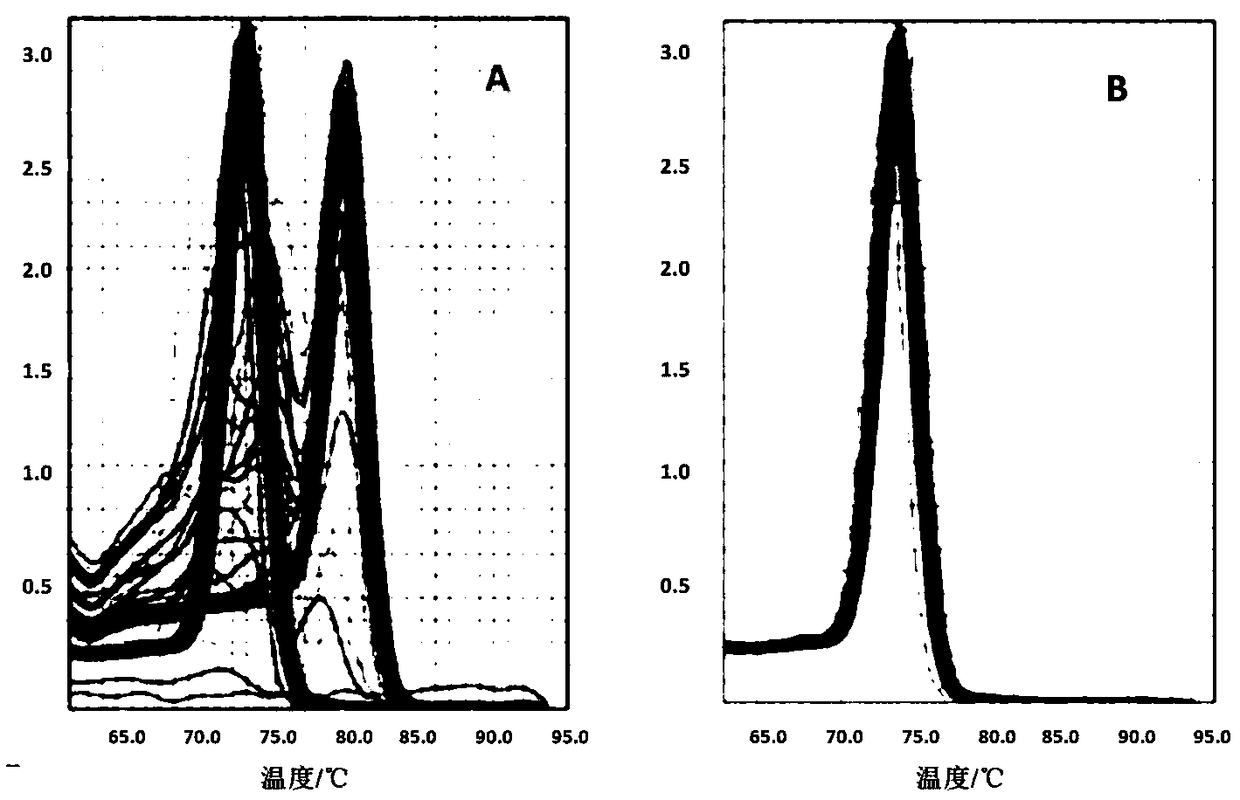
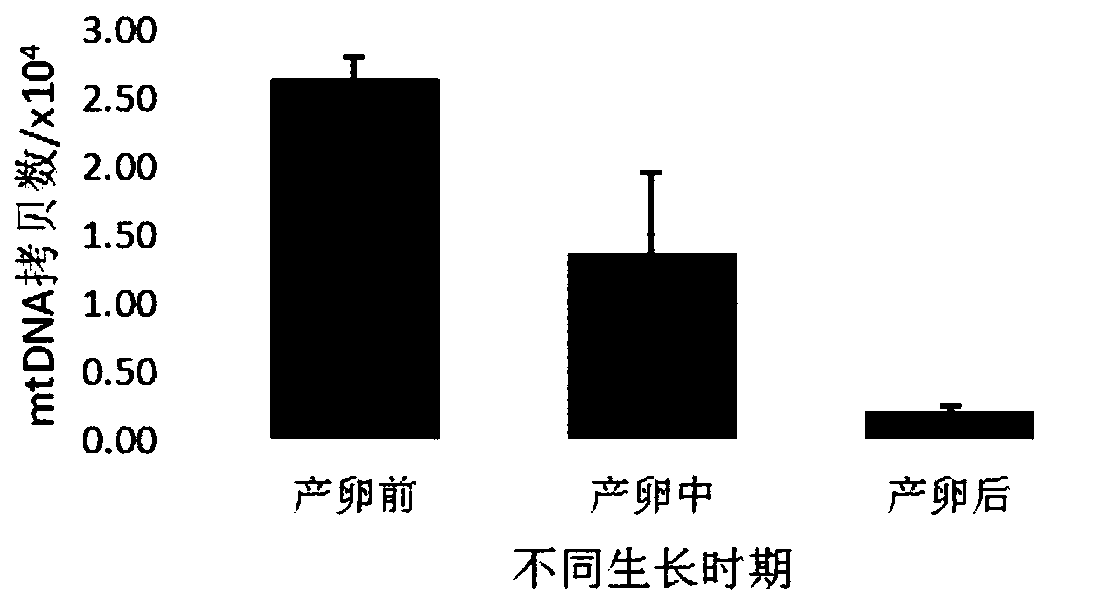
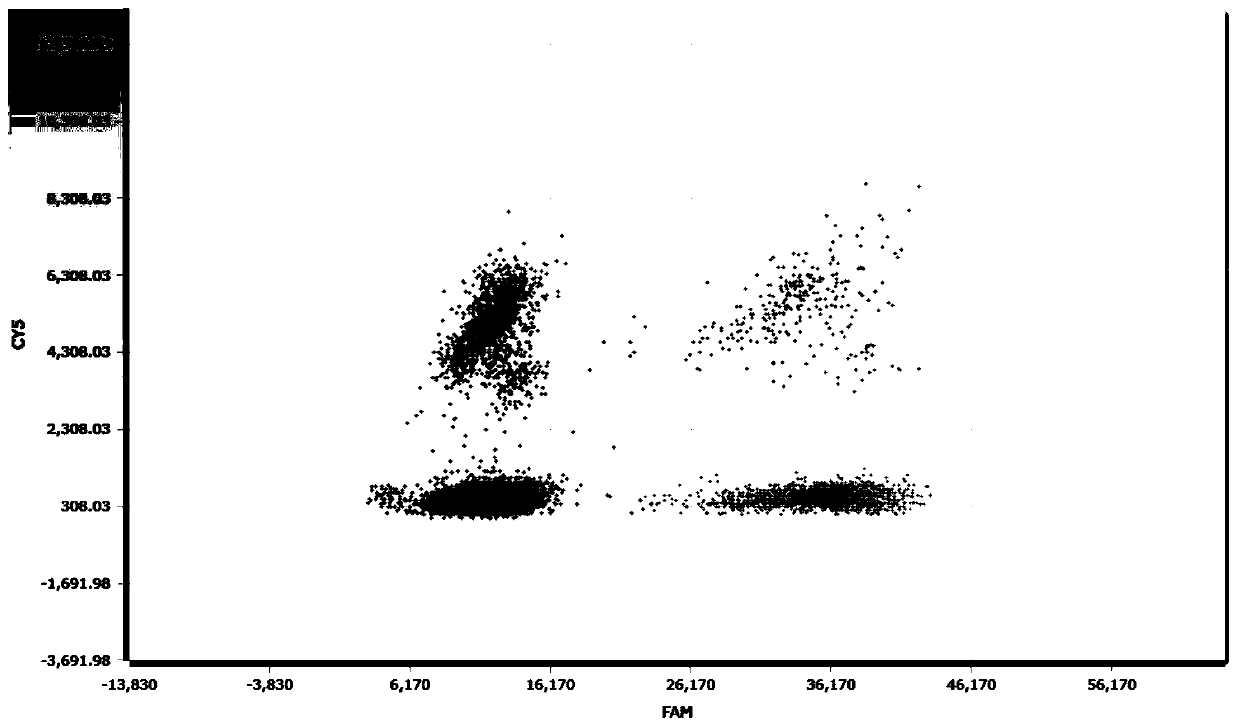
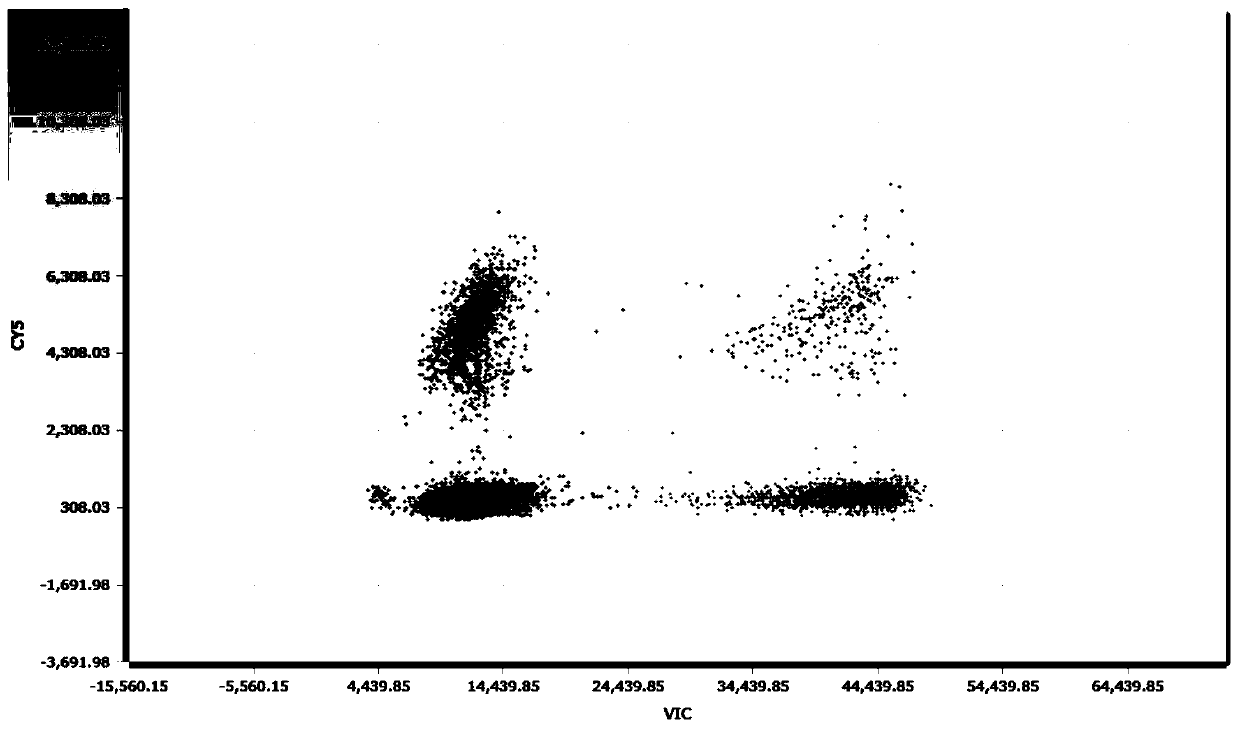
Change feature analysis method of liver mtDNA copy number in sepiella maindroni aging process
InactiveCN109486959AEasy to separateAvoid degradationMicrobiological testing/measurementDNA preparationPoly(butylene succinate)Standard Preparations
Owner:ZHEJIANG OCEAN UNIV
Short serial repeating sequence universal probe and design method and application thereof
PendingCN110343757ASolve the characteristicsSolve the costMicrobiological testing/measurementDNA/RNA fragmentationNon invasiveShort Tandem Repeat Profile
The invention relates to the field of in-vitro diagnosis, and more specifically relates to a universal probe for simultaneously detecting multiple sites of a genome, and a primer design method and anapplication kit thereof. The universal probe adopts a short tandem repeat sequence as a skeleton sequence, and solves the problems that the number of the probes in a reaction system in which the copynumber concentration is amplified is too large, and non-specific amplification is easily formed and the synthesis cost is high, and the concentration of the chromosome copy number can be proportionally amplified by designing multiple pairs of the primers on both sides of the universal probe position of the single copy gene fragment of the chromosome to-be-tested, which is especially suitable for the detection of certain very low concentration samples (such as whether the non-invasive detection of chromosome copy number generates variation or not).
Owner:领航医学科技(深圳)有限公司
DNA barcode preparation method and Phytophthora fragariae DNA barcode
ActiveCN105734049AEffective distinctionValid identificationMicrobiological testing/measurementDNA preparationDNA barcodingPhytophthora fragariae
The invention discloses a DNA barcode preparation method and a Phytophthora fragariae DNA barcode. The preparation method of the application comprises: sequencing target strain genome DNA by using high-throughput sequencing technology, subjecting sequencing results to bioinformatics analysis, analyzing an evolutional relation of gene fragments in the genome DNA by using a phylogenetic tree to obtain a target strain greater than or equal to 95% and less than 100% in similarity to strains of same genus, and subjecting single copy gene fragments not higher than 1000 bp to genetic distance analysis to obtain the target strain DNA barcode capable of genuinely reflecting genetic relations. The preparation method of the application uses the high-throughput sequencing technology to comprehensively analyze genome DNA and uses the phylogenetic tree and genetic distance analysis to obtain the DNA barcode; a single and effective way to prepare the DNA barcode is provided; the obtained DNA barcode is able to most effectively characterize target strains.
Owner:SHENZHEN CUSTOMS ANIMAL & PLANT INSPECTION & QUARANTINE TECH CENT
PCR method for detecting absolute telomere length
InactiveCN109943626ASuitable for high-throughput detectionReduce testing costsMicrobiological testing/measurementEscherichia coliFluorescence
The invention provides a PCR method for detecting absolute telomere length, and belongs to the field of molecular biological techniques and medical detection. According to the method, the absolute length of the telomere of the human can be conveniently and accurately detected. A gene synthesis method is adopted for synthesizing oligonucleotide sequences of the telomere and a single-copy gene 36B4respectively, the sequences are connected to a plasmid vector to construct a standard product, escherichia coli is utilized for in-vitro amplification of the plasmid standard product, the plasmid DNAis extracted, the concentration is measured, and the copy number and the total length of the telomere are calculated. The initial mass of the standard product is determined, gradient dilution is conducted, a fluorescence quantitative PCR method (Q-PCR) is adopted for obtaining standard curves of the telomere and the 36B4 separately, through the standard curves, the total length of the telomere andthe copy number of the 36B4 of the to-be-tested sample DNA are subjected to absolute quantification, and thus the average absolute telomere length of the to-be-tested sample DNA is calculated. The length of the telomere is detected through the absolute telomere length PCR method, and the PCR method has the advantages of being low in cost, short in period and high in accuracy, and is suitable forhigh-throughput detection, easy to clinically apply and popularize, and high in practicability.
Owner:山东荆卫生物科技有限公司
Meloidogyne molecular identification primer set, and preparation method and application thereof
InactiveCN103343169AAccurate identificationImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationNematostella
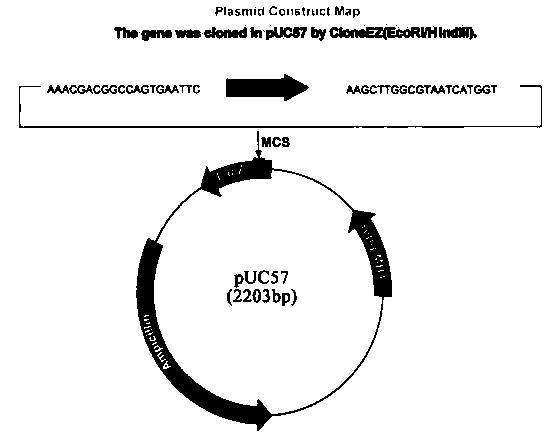
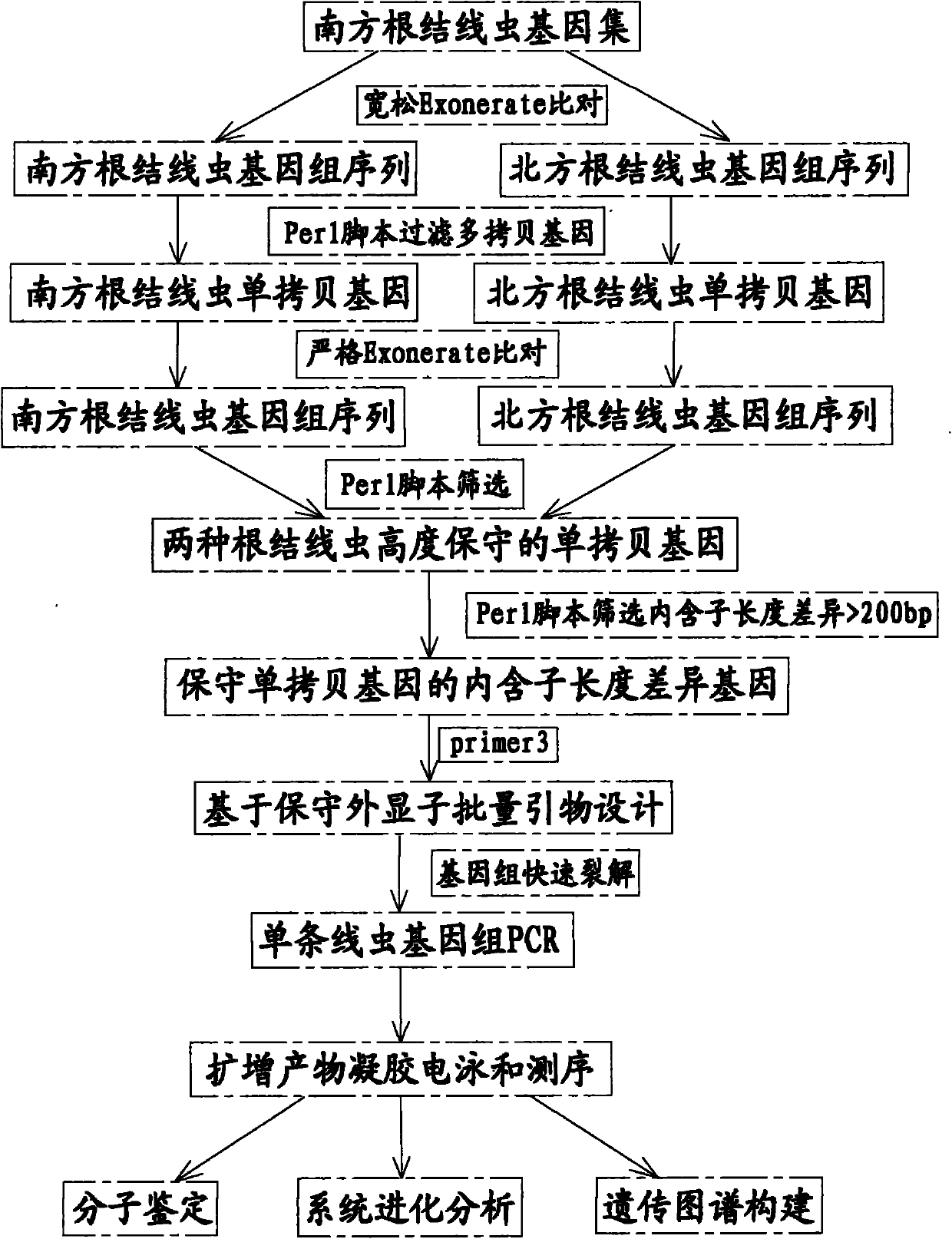
The invention provides a Meloidogyne molecular identification primer set which is prepared by the following steps: by comparing a predicted south Meloidogyne protein sequence respectively with south Meloidogyne genome and north Meloidogyne genome, setting the similarity of the comparison sequence, i.e. the percent parameter is 75, thereby obtaining the single-copy gene; by comparing the single-copy gene protein sequence respectively with the south Meloidogyne genome and the north Meloidogyne genome, setting the similarity of the comparison sequence, i.e. the percent parameter is 85, and screening the conserved single-copy gene in both of the two genomes; screening the gene of which the introne length difference between the two genomes is greater than 200bp; and designing primers on the basis of the exon sequences on two sides of the difference introne. Abundant primers are provided to enhance the detection accuracy; and the difference between the PCR (polymerase chain reaction) products of the primers via agarose gel electrophoresis is more than 200bp, and thus, the primers can be identified conveniently.
Owner:TOBACCO RES INST CHIN AGRI SCI ACAD
Detection method of vegetal single copy genes
InactiveCN102191325AThe detection process is shortHigh sensitivityMicrobiological testing/measurementFluorescence/phosphorescencePhysical MapsSingle copy
The invention relates to a detection method of vegetal single copy genes based on fluorescence in situ hybridization of quantum dots. By preparing quantum dot coupled DNA (deoxyribonucleic acid) as a probe, according to the traditional fluorescence in situ hybridization method, the hybridization detection of vegetal single copy genes is carried out on interphase nucleuses, chromosomes, DNA fibersor the like. In the method provided by the invention, a direct marking method is adopted, thus an antibody detection process is omitted; and because the quantum dots have high fluorescence intensity and can resist photo-bleaching, the sensitivity and resolution of hybridization are improved, and the vegetal single copy genes can be quickly and effectively located. The method provided by the invention can be widely used for drawing physical maps and analyzing the composition, structure, rearrangement, evolutionary relationship and the like of genomes.
Owner:WUHAN UNIV
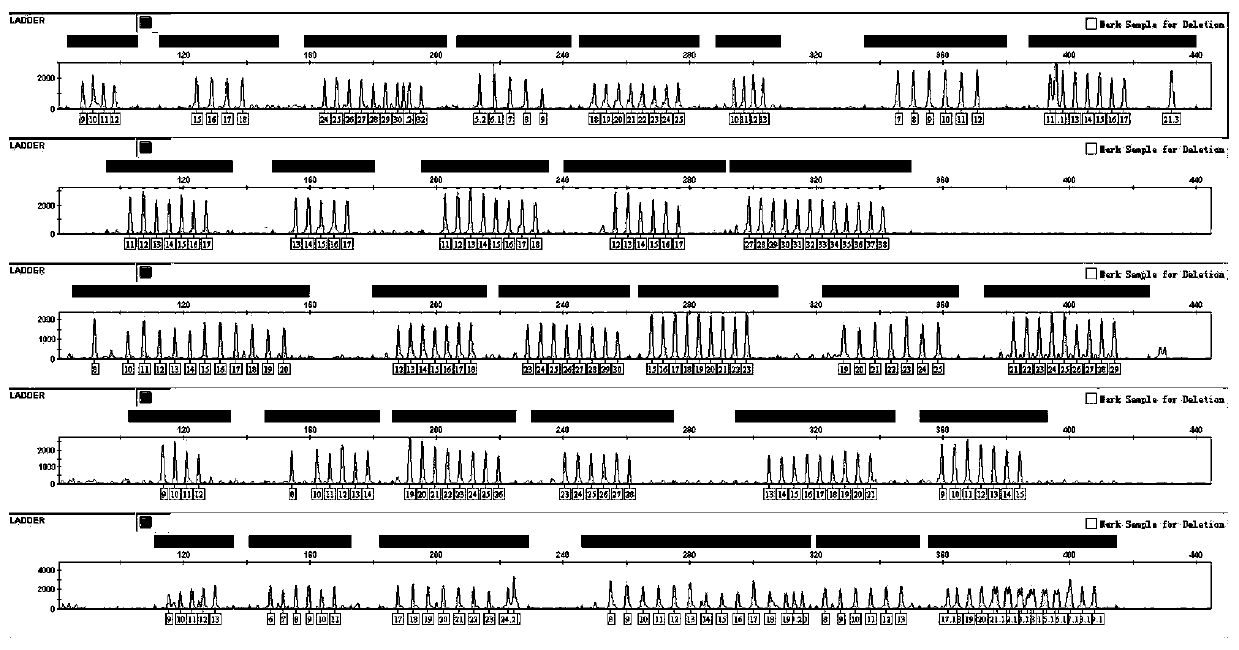
Composite amplification kit for simultaneously detecting 39 Y chromosome STR loci
InactiveCN110373475AHigh individual recognition rateImprove compatibilityMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceReaction system
The invention discloses a composite amplification kit for simultaneously detecting 39 human Y chromosome STR loci. 39 Y chromosome loci are simultaneously amplified in a composite amplification reaction system. The composite amplification kit comprises 26 single-copy loci, 3 double-copy loci, one three-copy loci and one four-copy loci. The 39 loci are divided into five groups through a loca arrangement mode which is unique in design and a specific primer sequence, and each group is labeled with a different fluorescent label. The kit has high resolution and high individual identification amongmale samples.
Owner:武汉瑞博科创生物技术有限公司 +1
A new type of endo-alginate lyase gene, engineering bacteria and application
ActiveCN105154458BIncrease enzyme activityHigh purityFungiMicroorganism based processesPichia pastorisAlginate lyase activity
The invention discloses a recombinant alginate lyase, its engineering bacteria and a method for preparing alginate oligosaccharides by hydrolysis. According to the newly isolated strain Vibrio sp.BZM‑1 from seaweed, the alginate gene-AlgM is screened; The expression vector of the gene, pHBM905BDM‑AlgM, is transformed into Pichia pastoris GS115 competent cells to obtain single-copy genetically engineered bacteria; construct, culture and verify multi-copy genetically engineered bacteria, efficiently express recombinant alginate lyase, and use this enzyme to hydrolyze alginic acid Sodium production of fucoidan oligosaccharides. GS115 / AlgM4 (CCTCC No: M2015500) with four copies of recombinant alginate lyase genetically engineered strain had a higher expression level. The activity of the recombinant alginate lyase of the present invention is as high as 31000U / mL.
Owner:BINZHOU MEDICAL COLLEGE
Quantitative test method of exopalaemon carinicaudamitochondrial genome copy number
InactiveCN106498044AIncrease concentrationQuick checkMicrobiological testing/measurementFluorescenceQuantitative PCR instrument
The invention discloses a quantitative test method of exopalaemon carinicauda mitochondrial genome copy number. The method comprises the steps of selecting the adenylyl transferase gene (ANT) and the ATP6 respectively as the single copy genes of the nuclear genome and the mitochondrial genome of white tailed shrimp, designing the primer and the fluorescent probe suitable for the real time quantitative PCR reaction of the ANT and ATP6 genes, preparing real time quantitative PCR reaction standard of the ANT and ATP6 genes, through drawing the standard curve (as shown in the attachment), and using the standard curve as the standard, and quantifying and calculating the copy number of the mitochondrial genome in the to-be-tested sample cells. The method can correctly and quantitatively test the mitochondrial genome copy number of the exopalaemon carinicauda tissue sample cells, and has the advantages of being high in specificity, and simple in operation. Merely the fluorescence quantitative PCR instrument is enough to complete the quantification of the exopalaemon carinicauda mitochondrial genome copy number.
Owner:HUAIHAI INST OF TECH
Ab initio generation of single copy genomic probes
ActiveUS20100240880A1Sugar derivativesSequence analysisReference genome sequenceRepetitive Sequences
Single copy sequences suitable for use as DNA probes can be defined by computational analysis of genomic sequences. The present invention provides an ab initio method for identification of single copy sequences for use as probes which obviates the need to compare genomic sequences with existing catalogs of repetitive sequences. By dividing a target reference sequence into a series of shorter contiguous sequence windows and comparing these sequences with the reference genome sequence, one can identify single copy sequences in a genome. Probes can then be designed and produced from these single copy intervals.
Owner:ROGAN PETER K
Single copy genomic hybridization probes and method of generating same
InactiveUS20090312533A1Eliminates spurious hybridizationSugar derivativesMaterial analysis by observing effect on chemical indicatorGenomic SegmentHybridization probe
Nucleic acid (e.g., DNA) hybridization probes are described which comprise a labeled, single copy nucleic acid which hybridizes to a deduced single copy sequence interval in target nucleic acid of known sequence. The probes, which are essentially free of repetitive sequences, can be used in hybridization analyses without adding repetitive sequence-blocking nucleic acids. This allows rapid and accurate detection of chromosomal abnormalities. The probes are preferably designed by first determining the sequence of at least one single copy interval in a target nucleic acid sequence, and developing corresponding hybridization probes which hybridize to at least a part of the deduced single copy sequence. In practice, the sequences of the target and of known genomic repetitive sequence representatives are compared in order to deduce locations of the single copy sequence intervals. The single copy probes can be developed by any variety of methods, such as PCR amplification, restriction or exonuclease digestion of purified genomic fragments, or direct synthesis of DNA sequences. This is followed by labeling of the probes and hybridization to a target sequence.
Owner:CHILDRENS MERCY HOSPITAL
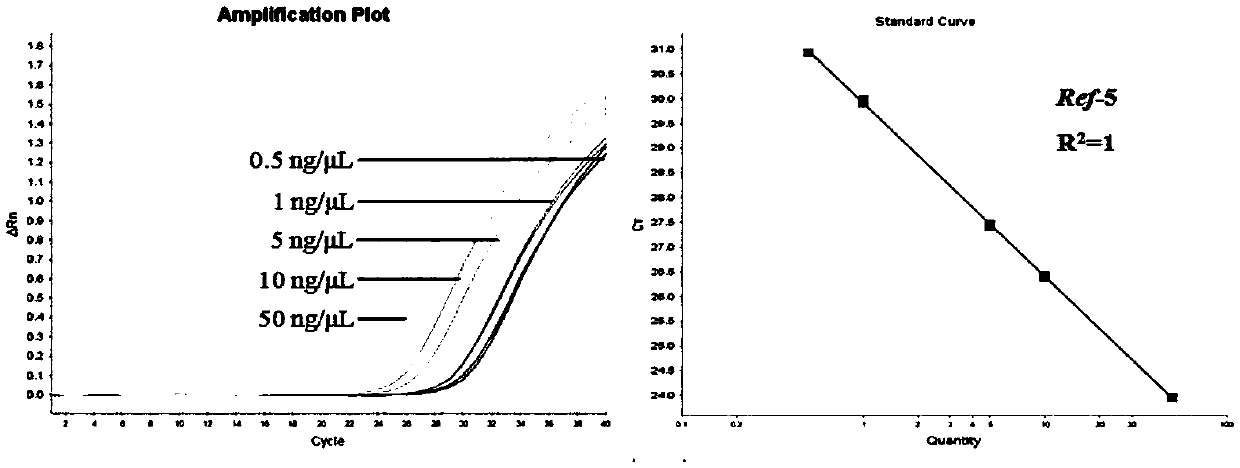
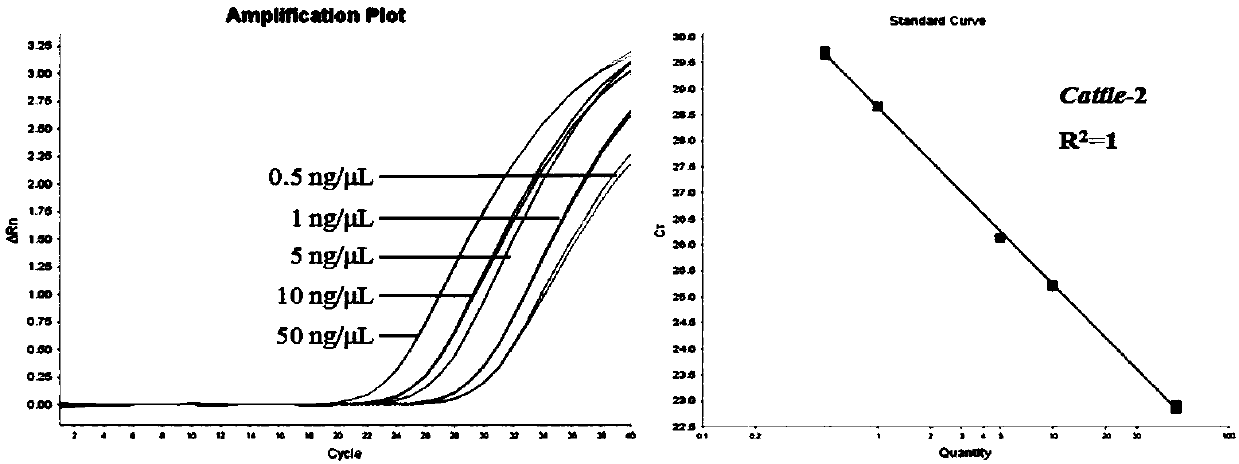
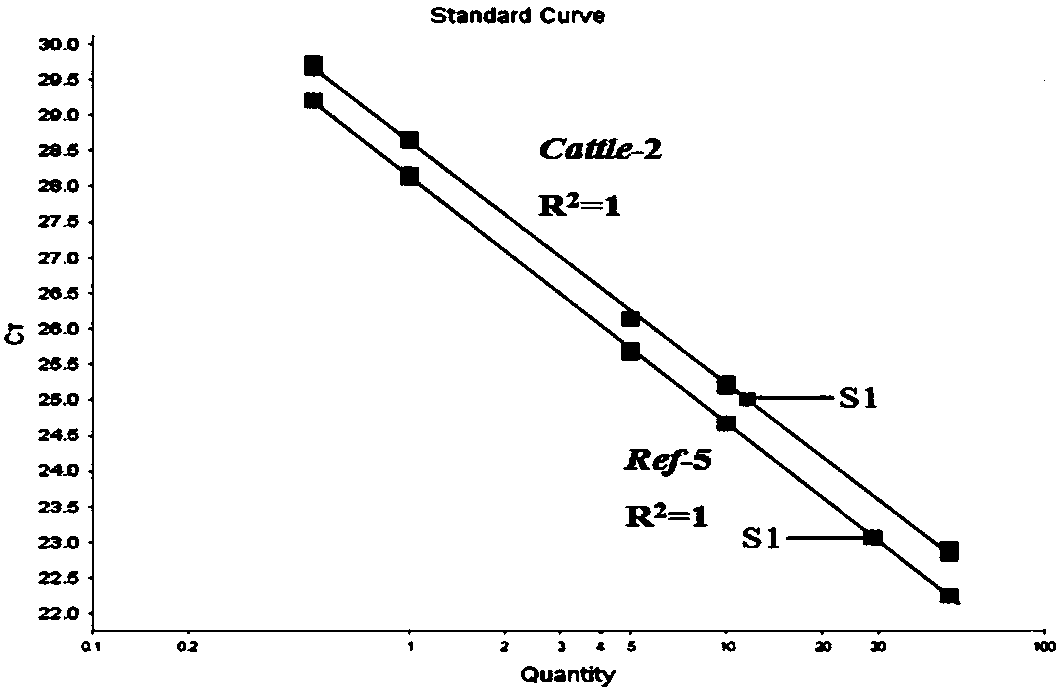
Method for quantitatively detecting bovine-derived materials in livestock and poultry meat based on QPCR (quantitative polymerase chain reaction)
The invention discloses a method for quantitatively detecting bovine-derived materials in livestock and poultry meat based on QPCR (quantitative polymerase chain reaction). By the method, the contentof bovine-derived materials in meat products is quantitatively analyzed by real-time fluorescent PCR technology; 15 animal genome sequences are analyzed by applying a bioinformatical method; universalprimers and specific primers are designed by screening single-copy genes; the percentage of bovine-derived species specific genes is corrected by an internal reference gene; and the deviation of quantity value results is eliminated, thereby obtaining the authentic and reliable food adulteration value, and effectively improving the sensitivity and accuracy of the detection method. The detection method disclosed by the invention can complete the identification work of the bovine-derived materials within 3 hours, has the characteristics of simple and easy operation, high sensitivity, rapidity, low cost, convenient promotion, and the like, and opens up a broad prospect for quantized adulteration identification of animal-derived materials.
Owner:TIANJIN INSTITUE OF QUALITY STANDARD & TESTING OF AGRICULTUAL PRODS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com