Meloidogyne molecular identification primer set, and preparation method and application thereof
A root-knot nematode and molecular identification technology, applied in the fields of biotechnology and genetic engineering, can solve the problems of long time-consuming, insensitive, complicated operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] The genome sequencing of Meloidogyne incognita and Meloidogyne incognita have been completed by French and American scientists respectively. Genomic sequence data and predictions can be downloaded from the genome databases of Meloidogyne infra (http: / / www.inra.fr / meloidogyne_incognita) and Meloidogyne (http: / / www.pngg.org / cbnp / index.php) Gene and protein sequence data.
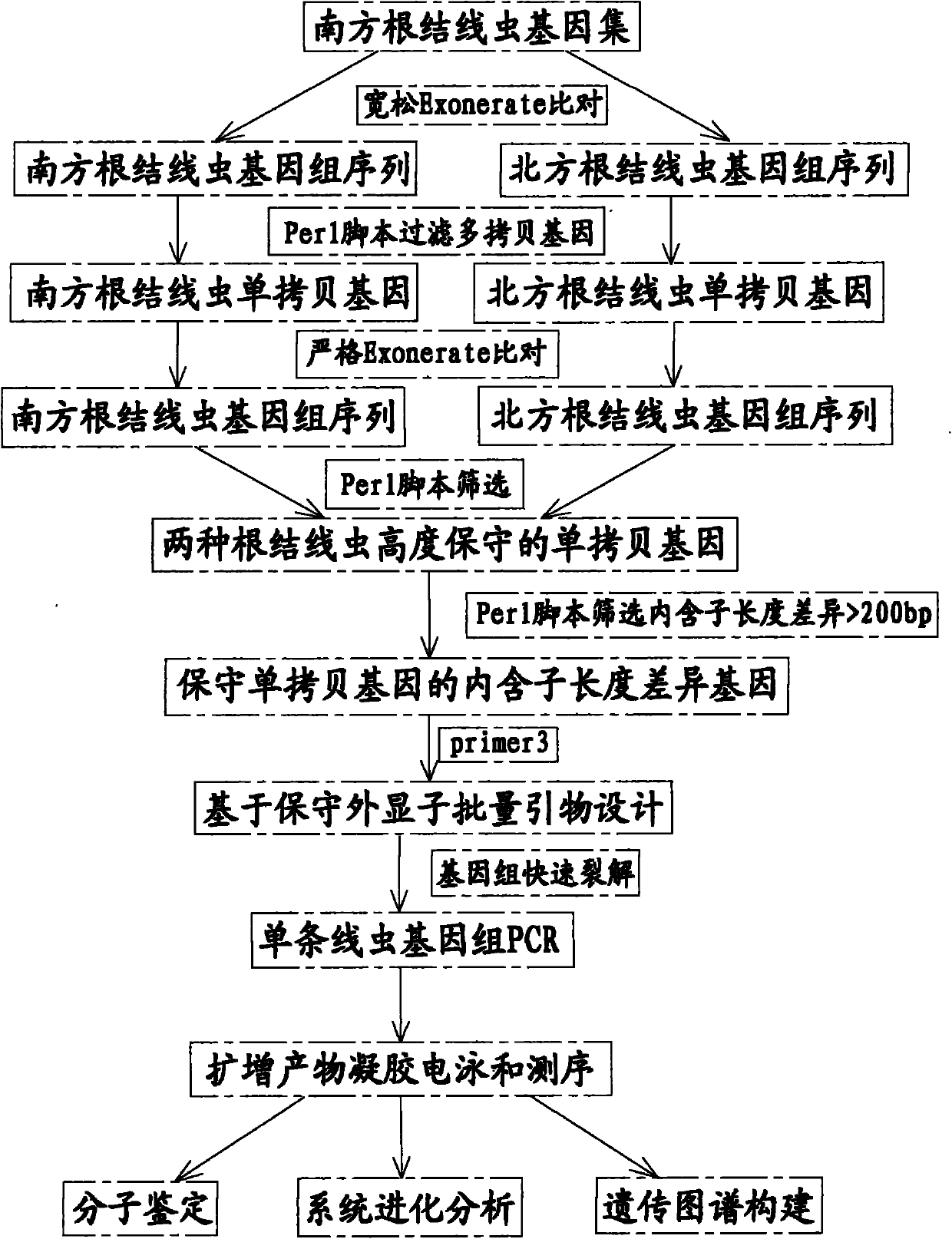
[0070] a. The sequence comparison software Exonerate can be used to align the predicted protein sequences of Meloidogyne incognita with Meloidogyne incognita and Meloidogyne incognita genomes respectively, and set the similarity of the aligned sequences, that is, the percent parameter to 75. The similarity setting in this step is relatively loose, filtering multiple copies of homologous gene families as much as possible to obtain single copy genes. The Perl program was used to screen single-copy genes from the comparison results, and the sequence information of single-copy genes in the two genomes was ext...
Embodiment 2
[0079] One of the primer pairs in Example 1 was used to identify root knot nematodes in the sample.
[0080] (1) Collect samples of root knot nematodes in the south, north and Florida. Wash the nematodes in ddH2O, pick a single nematode and put it into a 200μL PCR tube (containing 8μL ddH 2 O and 1 μL 10X PCR buffer), place in liquid nitrogen for 1 minute, heat at 85°C for 2 minutes, add 1 μL of 1 mg / mL proteinase K to the PCR tube, heat at 56°C for 15 minutes, and heat at 95°C for 10 minutes to obtain a nuclear genomic DNA extract.
[0081] (2) Amplify the obtained nuclear genomic DNA extract directly, and the PCR primer pair used is WBMinc00222:
[0082] Upstream primer WBMinc00222.L: tcataacatcaatagaggat
[0083] And the downstream primer WBMinc00222.R: cgaaacaatgctgccgtaca;
[0084] The PCR reaction system is: 10XPCR buffer (no Mg 2+ ) 5ul, 25mmol / L MgCl 2 5ul, 0.1mmol / L dNTP4ul, 10umol / L upstream primer and downstream primer, 5U / uL Tag enzyme 0.6uL, plus DNA extract to supplement ...
Embodiment 3
[0088] One of the primer pairs in Example 1 was used to identify root knot nematodes in the sample.
[0089] (1) Collect samples of root knot nematodes in the south, north and Florida. Put the nematode into ddH 2 O wash, pick a single nematode and put it in a 200μL PCR tube (containing 8μL ddH 2 O and 1 μL 10X PCR buffer), place in liquid nitrogen for 1 minute, heat at 85°C for 2 minutes, add 1 μL of 1 mg / mL proteinase K to the PCR tube, heat at 56°C for 15 minutes, and heat at 95°C for 10 minutes to obtain a nuclear genomic DNA extract.
[0090] (2) Amplify the obtained nuclear genomic DNA extract directly, using PCR primer pair as WBMinc00818:
[0091] Upstream primer WBMinc00818.L: aatgttattggaactaattt
[0092] And the downstream primer WBMinc00818.R: tccatagcacgcattgcttg;
[0093] The PCR reaction system is: 10XPCR buffer (no Mg 2+ ) 5ul, 25mmol / L MgCl 2 5ul, 0.1mmol / L dNTP4ul, 10umol / L upstream primer and downstream primer, 5U / uL Tag enzyme 0.6uL, plus DNA extract to supplement dd...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com