A new type of endo-alginate lyase gene, engineering bacteria and application
A technology of alginate lyase and engineering bacteria, applied in the field of gene sequences of novel endo-type alginate lyase AlgM, can solve problems such as low substrate concentration, inability to meet production requirements, low hydrolysis efficiency, etc., and achieve production cost low effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] 1. The first step of the inventive method is to screen the alginate lyase AlgM gene:
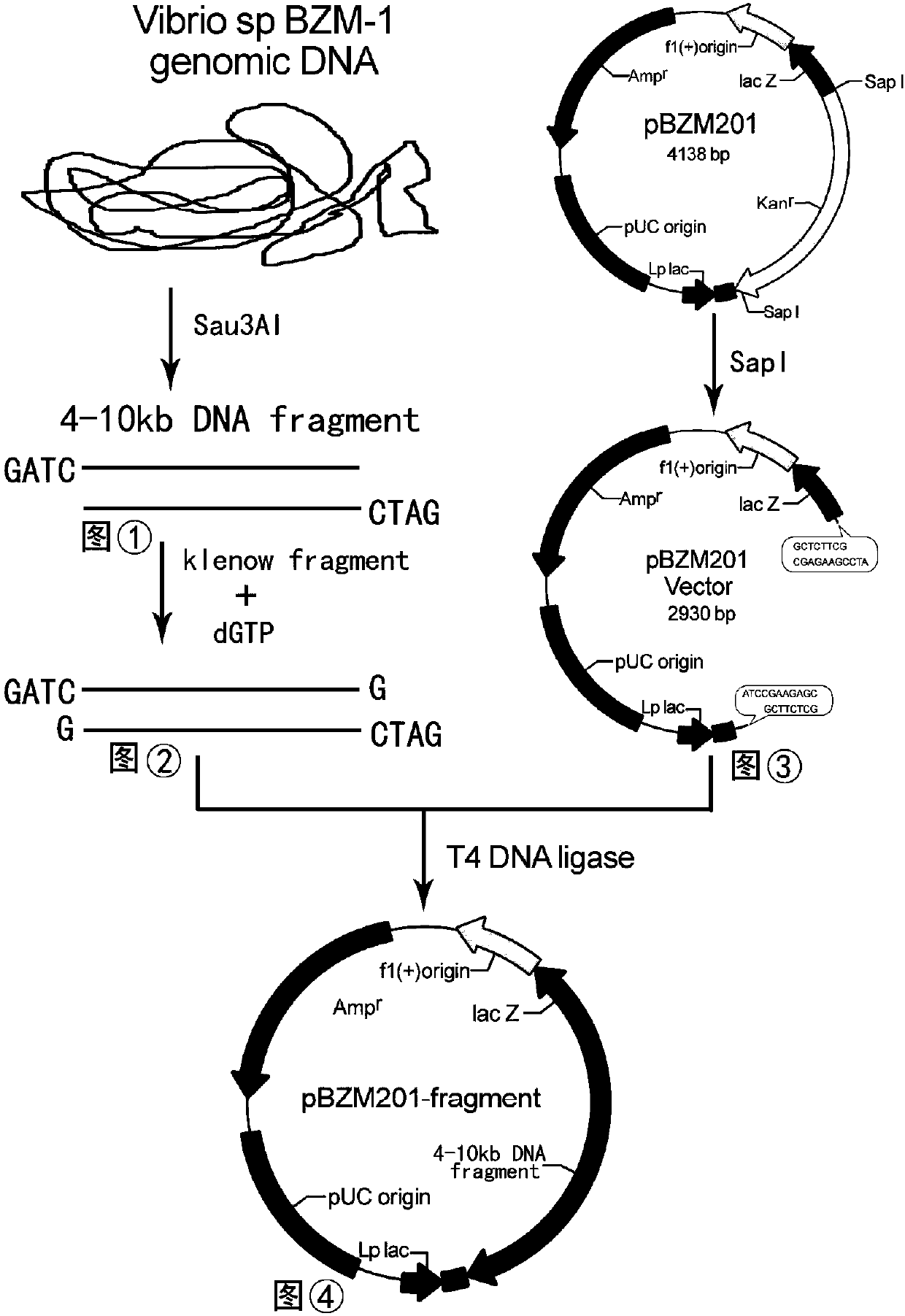
[0065] According to the newly isolated strain Vibrio sp.BZM-1 from alginate lyase AlgM, the genome library of the bacteria was established. First, the genomic DNA of the bacteria was extracted, partially digested with the restriction endonuclease Sau3A I, and recovered from the agarose gel. For 4-10kb fragments, these fragments were supplemented with dGTP, and the 3-7kb fragments were recovered by regelation. At the same time, the vector pBZM201 was linearized by restriction endonuclease Sap I, and the fragments were recovered as vectors, and the complementary base 4- The 10kb fragment was ligated with the pBZM201 vector linearized by Sap I digestion at 16°C for 24h, and then transformed into Escherichia coli Xl10-gold competent cells to obtain the genome library of the bacterium (see figure 1 ). Cultivate on the LB medium plate containing sodium alginate and ampicillin for 20 hours...
Embodiment 2
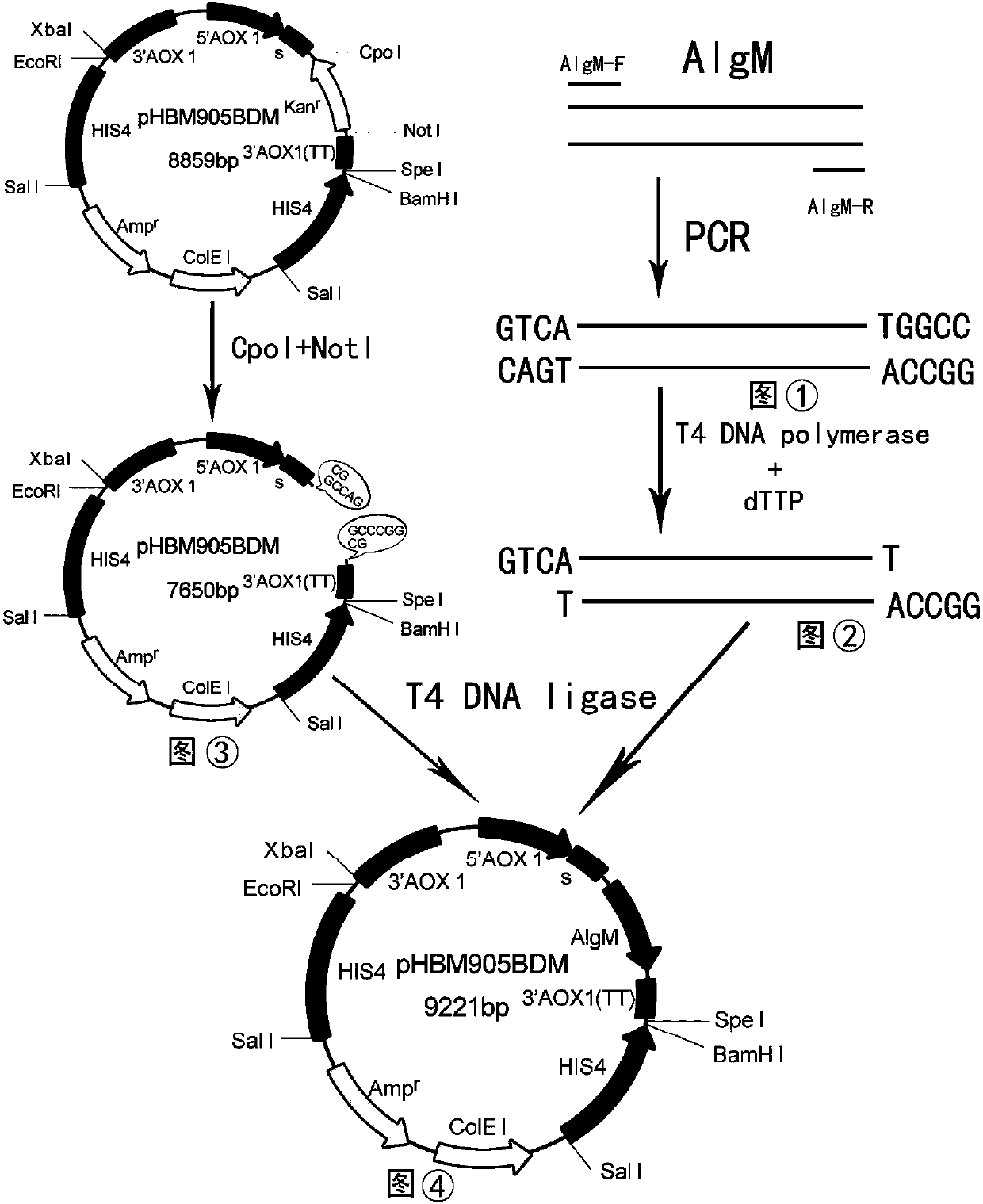
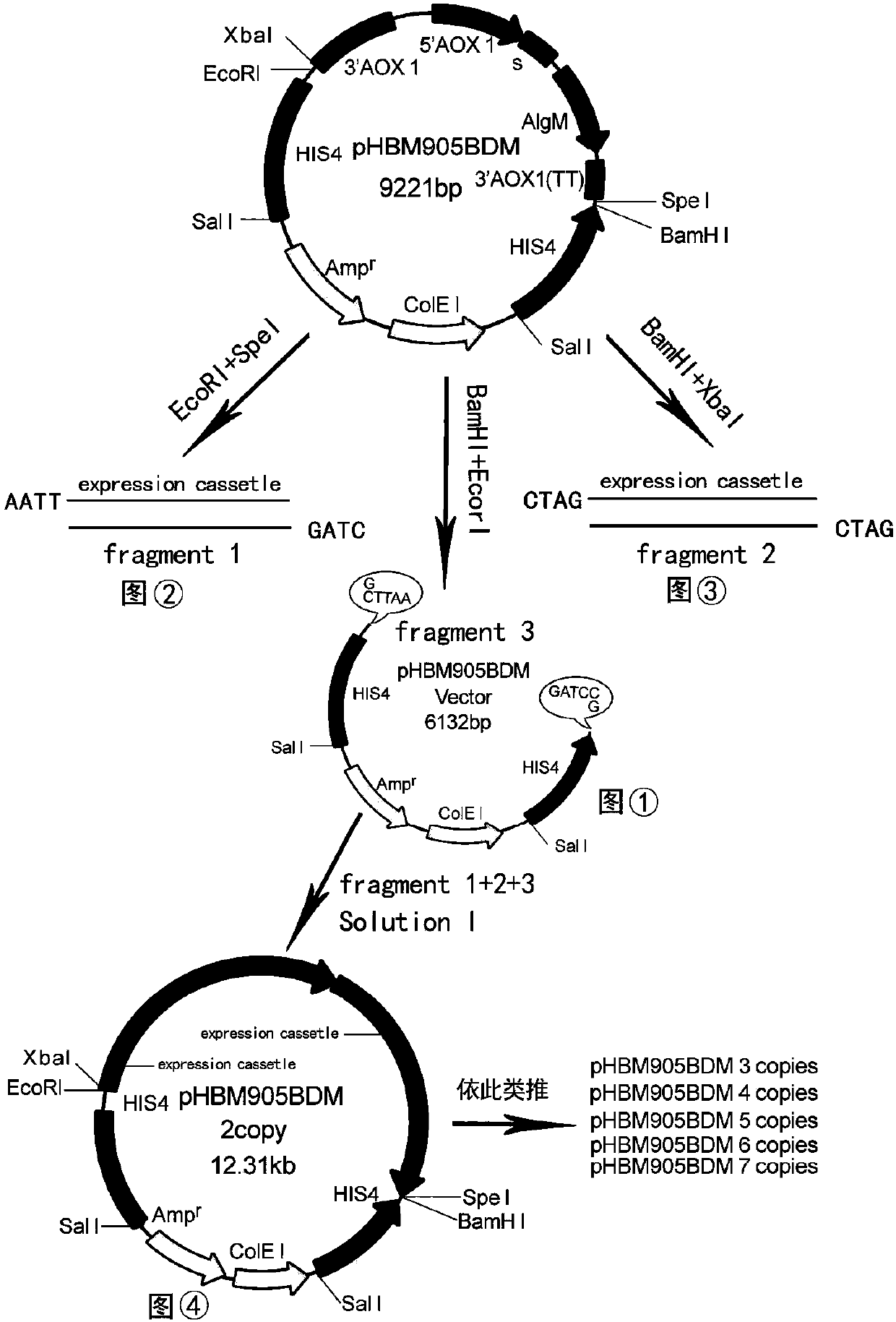
[0071] Inoculate Pichia pastoris strains with three-copy, four-copy, five-copy, six-copy and seven-copy expression cassette structures in 100ml BMGY medium respectively, and culture them at 28°C-30°C, 260rpm for 40-50h until OD 600 20-30, centrifuge at room temperature at 5000rpm for 5min, and collect the bacteria; transfer the bacteria to 100ml BMMY medium, culture at 28°C-30°C, 260rpm, and add methanol every 12h (addition amount is the medium volume 1.0%), after induced expression, the supernatant at the time of the highest expression was taken for SDS-PAGE electrophoresis to analyze the protein expression of Pichia strains with different copy expression cassette structures, three copies, four copies and five copies of expression The expression time of Pichia pastoris strains with cassette structure was the supernatant of 96h, 120h and 144h. Take 8 microliters of supernatant from each sample for SDS-PAGE electrophoresis. It was found that at 144h, four copies of Pichia pastor...
Embodiment 3
[0073] Using the recombinant alginate lyase crude enzyme liquid obtained in the shake flask in Example 2, hydrolyzing sodium alginate with different concentrations to prepare alginate oligosaccharides.
[0074] Raw material: sodium alginate
[0075] The concentrations of the hydrolyzed substrates were respectively prepared as 10%, 15%, 20%, 25%, and 30%; 100ml of alkaline water with a temperature of 30-40°C and a pH between 8 and 9 was used as a solvent, and an appropriate amount of sodium alginate was added. Stir and prepare 100ml of reaction solutions with substrate concentrations of 10%, 15%, 20%, 25%, and 30%, respectively, and then add 1000ul, 1500ul, 2000ul, 2500ul, 3000ul of crude enzyme solution, adjust the pH to 8-9. The hydrolysis time was 16, 14, 12, 10, 8 hours respectively. The reaction solution is centrifuged to remove residues, the supernatant is filtered, ultrafiltered, and vacuum spray-dried to obtain the finished product of fucoidan oligosaccharide. The ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com