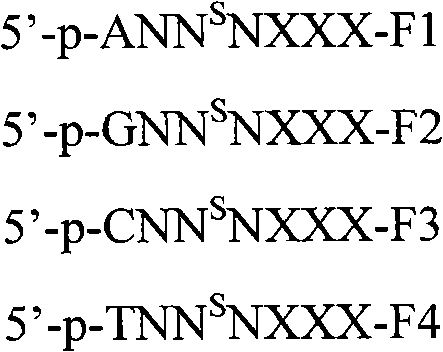Application of oligonucleotide probe containing dithio nucleotide on detecting DNA sequences
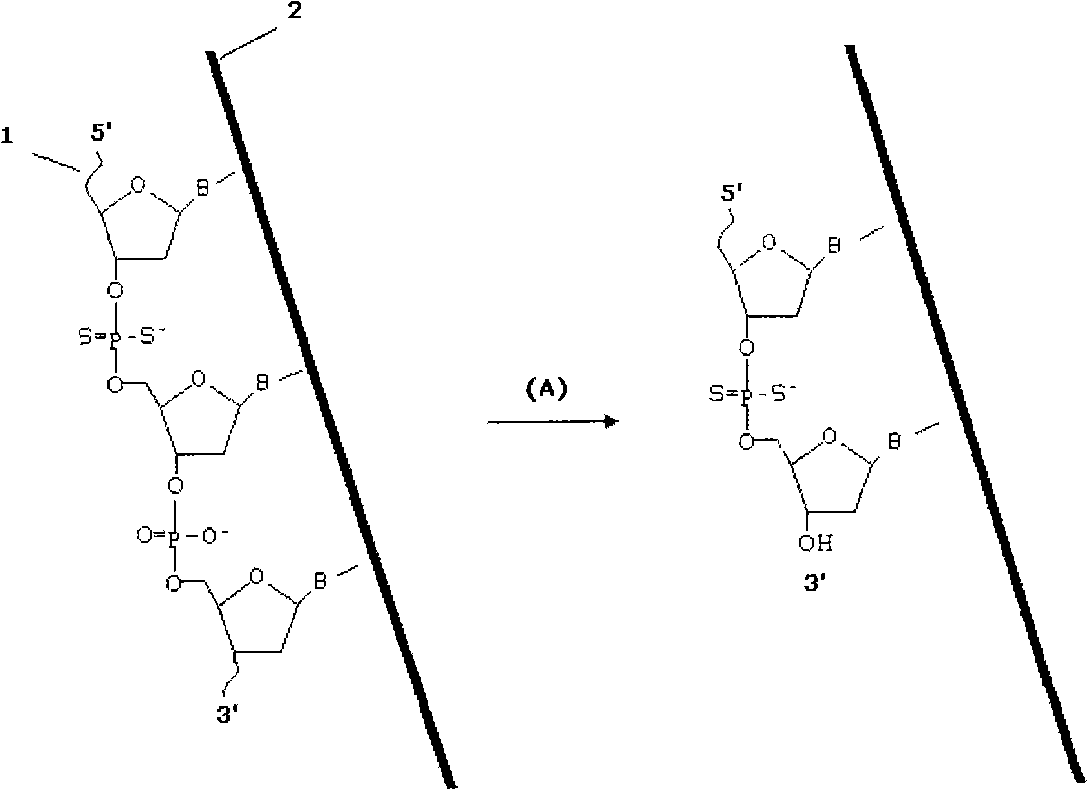
The technology of dithionucleoside oligonucleotide and dithionucleoside is applied to DNA sequencing probe and its application field in determining DNA sequencing sequence, and can solve the problem of non-degrading single-stranded DNA double-stranded DNA performance, It can not cut the phosphodiester bond, does not degrade the phosphodiester bond, etc., to achieve the effect of accurate and reliable measurement, increased reading length, and accurate cutting position
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
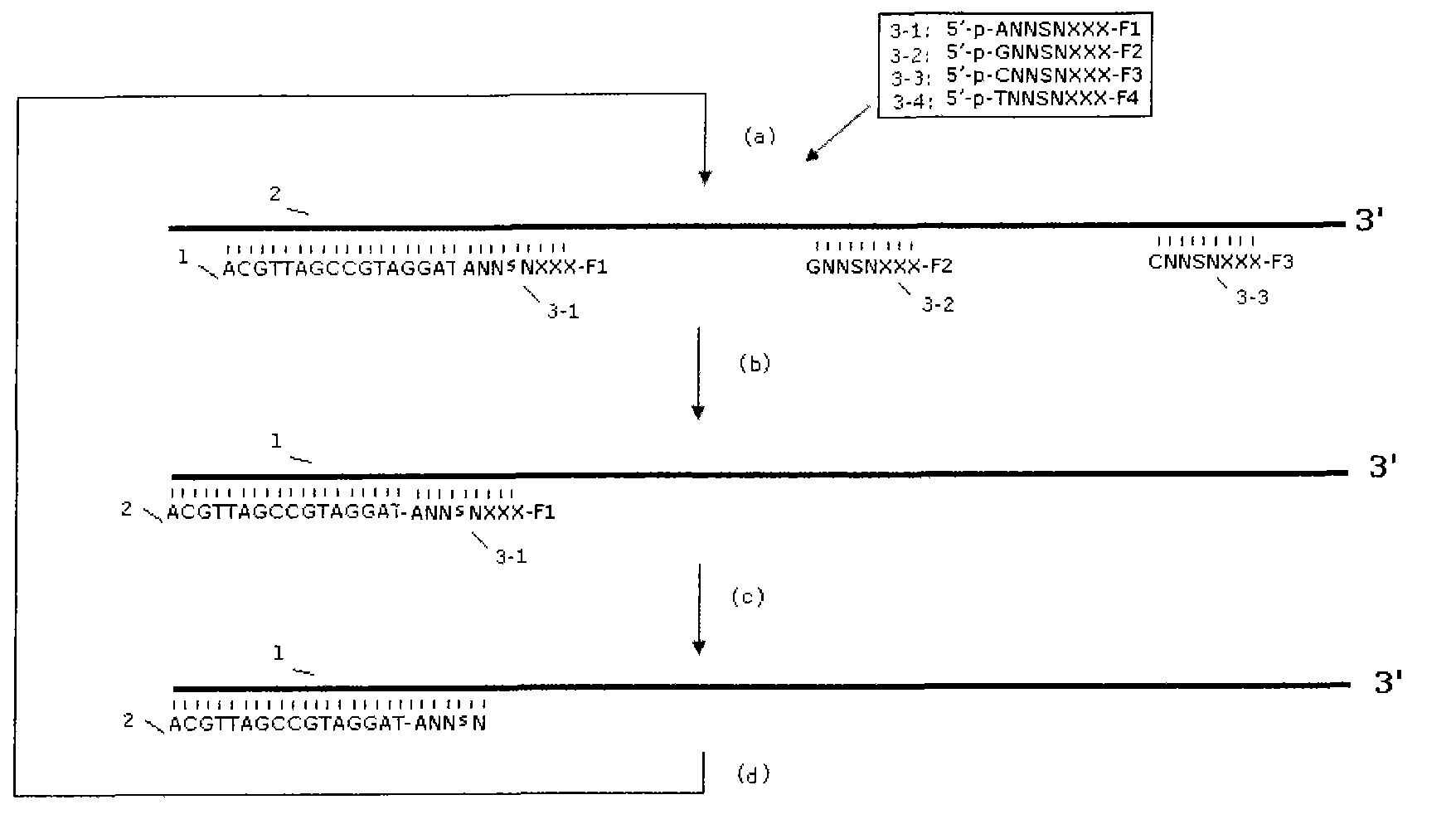
Image
Examples
Embodiment 1
[0053] Hybridization-ligation fluorescent-labeled sequencing assays encompass the entire human genome.
[0054] Cut the human genome with enzymes (or sonicate) into fragments with a size of 50-1000 bases, and use a pair of universal linkers (assumed to be 20 bases) to these fragmented nucleic acid sequences under the action of ligase T4 ligase (Shanghai Bioengineering Co., Ltd.) was used for ligation at 37° C. for 2 hours. The complementary oligonucleotide sequence of the linker is used as the amplification primer (one of which is pre-immobilized on the microbeads: the immobilization method can be a biotin-modified oligonucleotide synthesized by Yingjun (Shanghai) Bioengineering Co., Ltd. and New England Biolabs (USA) Co., Ltd. The produced streptavidin-modified microbeads were reacted at room temperature for 1 hour), and the sequence of one of the linker oligonucleotides was exactly the same as that of the sequencing primer in the subsequent sequencing reaction
[0055] The ...
example 2
[0072] Sequencing of three known DNA template sequences
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com