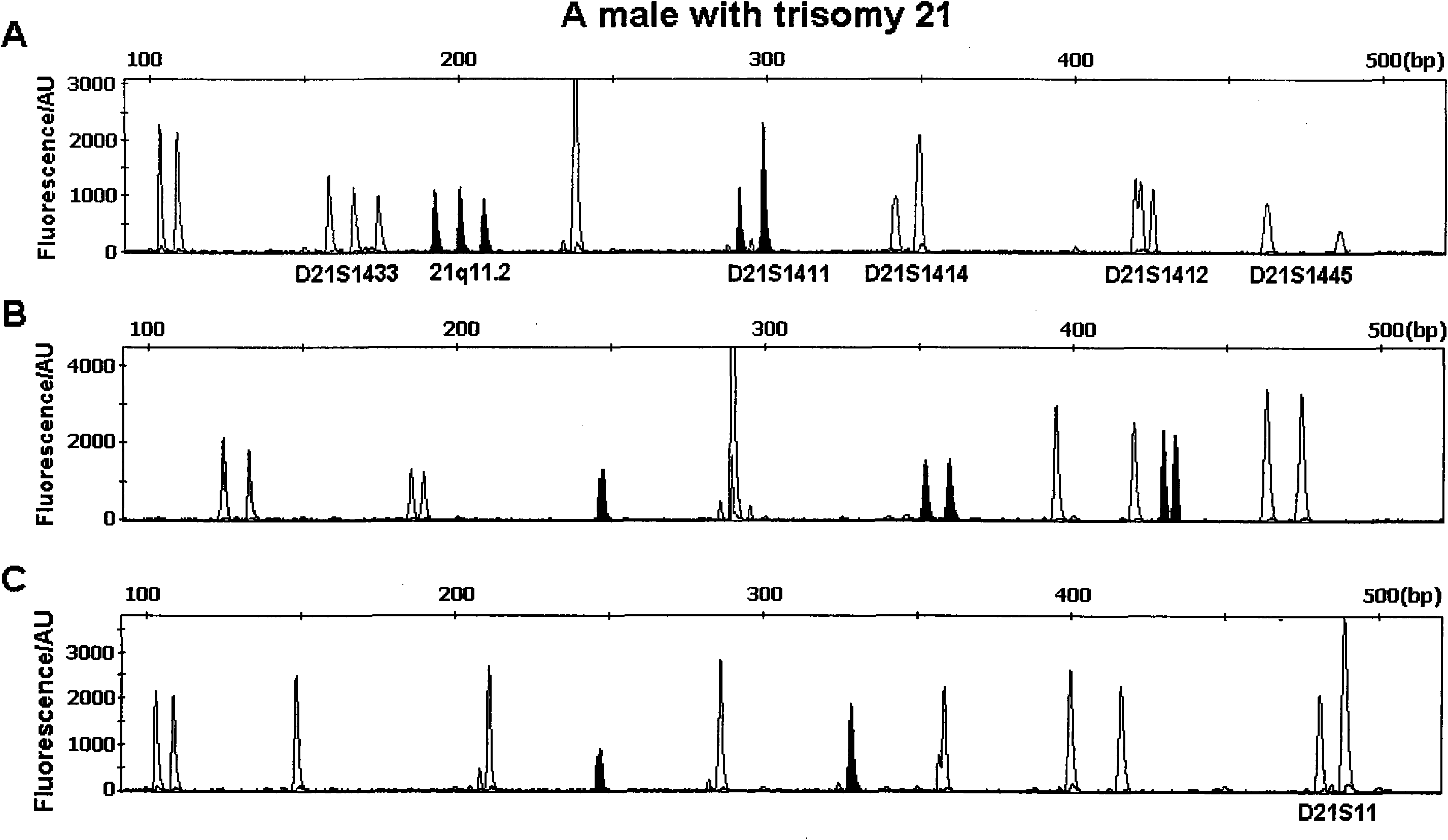
Multi-QF-PCR STR detection system for performing fast diagnosis on numerical abnormality of chromosomes
A QF-PCR STR and chromosome number technology, applied in the field of QF-PCR rapid diagnosis, can solve the problems of low heterozygosity rate of STR loci, poor reproducibility, insufficient STR loci, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment one: detection method of the present invention
[0036] Specimens for prenatal diagnosis include amniotic fluid, villi, fetal blood, etc. Whole blood samples need ACD anticoagulation, amniotic fluid samples need to be centrifuged at 3000rpm to collect cells, and the villi are soaked in normal saline. Various specimens were stored at 4°C, and DNA was extracted within 48 hours. DNA was extracted using a kit, such as QIAamp DNA Mini kit (QiaGen), and operated strictly according to the instructions. Finally, the DNA product was dissolved in double-distilled water, and the concentration and purity were measured on the N.D1000V3.7 DNA analyzer, and the DNA concentration was adjusted to 10-50ng / μL. OD260 / 280 ratio between 1.6-2.1 is a qualified DNA sample.
[0037] PCR reaction: DNA polymerase using Takara Taq TM For Hot Start Version, the PCR reaction uses a 25μL system, and the amount of DNA template is 15-70ng. Three sets of reactions A, B, and C were perfo...
Embodiment 2
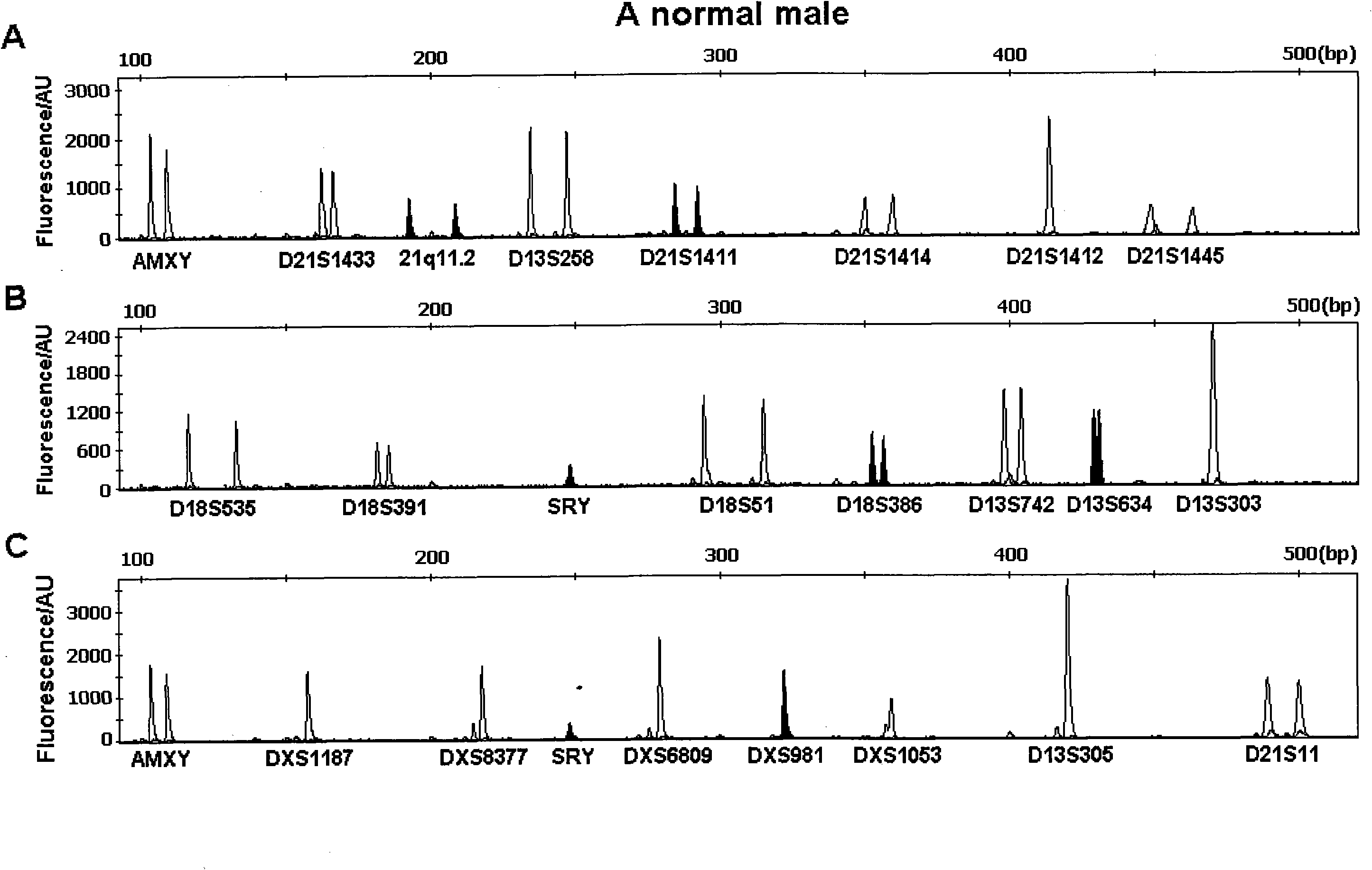
[0046] Embodiment 2: QF-PCR detection of a healthy male
[0047] refer to figure 1 As shown, using the method of Example 1, DNA was extracted from 200 μL of a peripheral blood sample of a healthy adult male with a normal karyotype confirmed by karyotype analysis, and a total of 45 μL of DNA with a concentration of 23.5 ng / μL was obtained, and the OD260 / 280 ratio was 1.82. For qualified DNA samples. Take 2 μL of DNA each for PCR reactions and fragment analysis in groups A, B, and C. The analysis results of the collected fluorescence data by GeneMapper software are shown in Table 4:
[0048] Table 4 GeneMapper software to the data analysis results of the QF-PCR fluorescence signal of a healthy male
[0049]
[0050]
[0051] The fluorescence signal intensity of all allele peaks in this sample is between 500-8000, and the fluorescence signal intensity is suitable for karyotype judgment; the gender determination gene AMXY shows a 1:1 (AMX:AMY) double peak, and the male-sp...
Embodiment 3
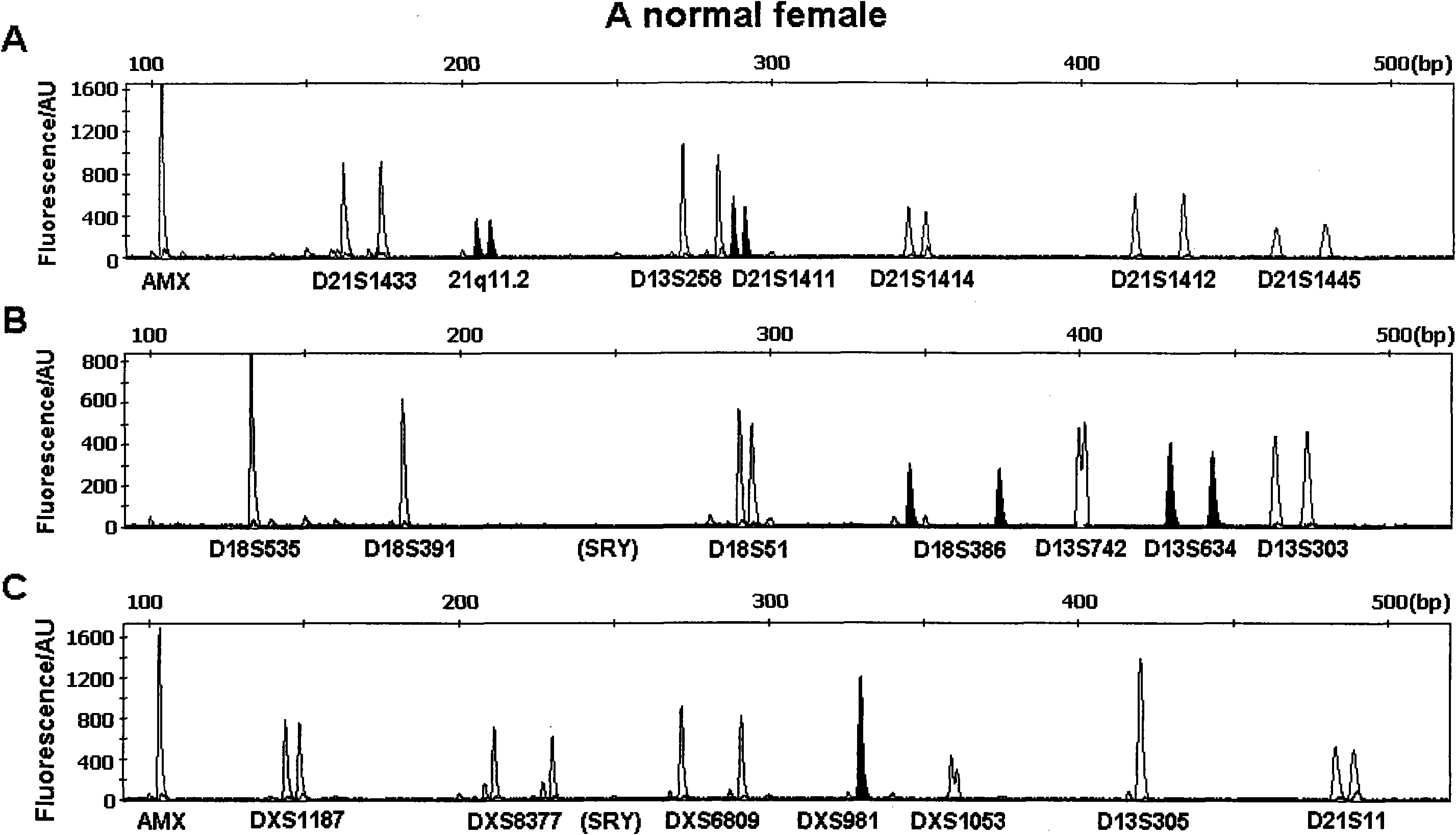
[0052] Embodiment three: the QF-PCR detection of a healthy female
[0053] refer to figure 2 As shown, using the method of Example 1, DNA was extracted from 200 μL of a peripheral blood sample of a healthy adult female confirmed by karyotype analysis as a normal karyotype, and a total of 45 μL of DNA with a concentration of 26 ng / μL was obtained, and the OD260 / 280 ratio was 1.91, which was Qualified DNA samples. Take 2 μL of DNA each for PCR reactions and fragment analysis in groups A, B, and C. The analysis results of the collected fluorescence data by GeneMapper software are shown in Table 5:
[0054] Table 5 GeneMapper software to the data analysis results of the QF-PCR fluorescence signal of a healthy female
[0055]
[0056]
[0057] *DXS1053 site PCR amplified fragment has a simple repetition of two bases "CA" in the middle, resulting in slippage in the PCR process, so the product doublet ratio is not strictly close to 1:1 (<1.6), which is still normal.
[005...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com