Primer for detection of shigella and detection method
A Shigella, primer amplification technology, applied in microorganism-based methods, biochemical equipment and methods, and microbial assay/inspection, etc., can solve problems such as low specificity of target sequence amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030]To detect Shigella using loop-mediated isothermal gene amplification, follow the steps below:
[0031] (1) Pretreatment of samples to be tested
[0032] Suspend the tested sample in sterile normal saline, centrifuge at 10,000r / min for 3min, remove the supernatant, then wash the cells twice with sterile normal saline, and air-dry;
[0033] (2) Preparation of template
[0034] Add 50 μL ddH to the cell sample obtained above 2 O. After fully mixing, place in a boiling water bath at 100°C for 10 minutes, in an ice bath for 5 minutes, and centrifuge at 12,000 r / min at 4°C for 5 minutes. The supernatant is the DNA template.
[0035] (3) Loop-mediated isothermal amplification (LAMP) reaction
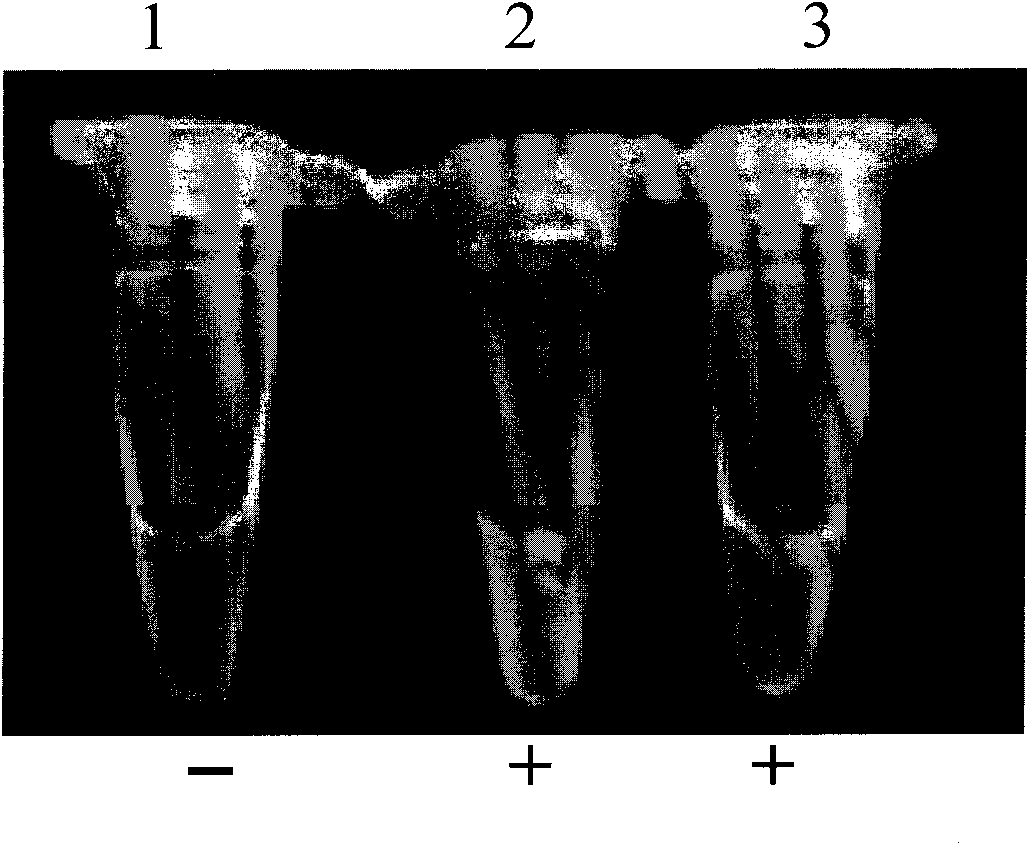
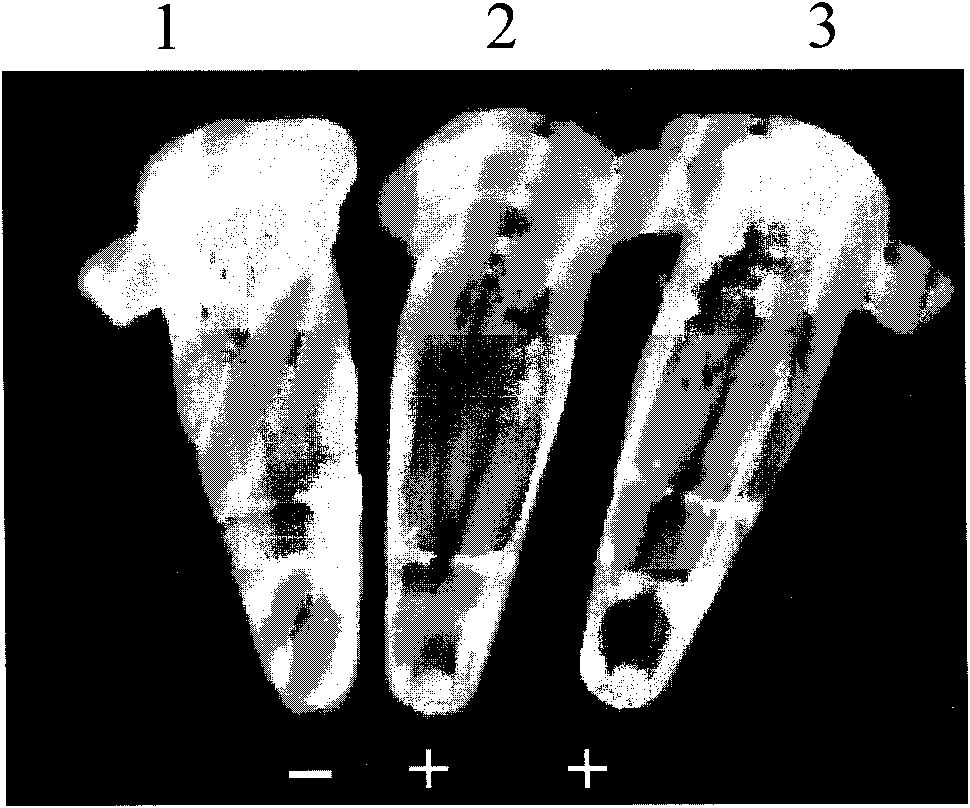
[0036] Take 2 μL of the above template, 1 μL of each primer, 1 μL (8U) of Bst DNA polymerase, 7.5-12.5 μL of the amplification reaction solution, add water to the reaction system of 25 μL, and react at a constant temperature of 65 °C for 1.5 h; then place it at 80 °C for 10 min, stop ...
Embodiment 2
[0042] To detect Shigella from food samples, proceed as follows:
[0043] (1) Pretreatment of samples to be tested
[0044] Take 25g of the sample and add it to 225mL of GN enrichment solution, and homogenize it in a tissue grinder to make a homogenate. The homogenate was diluted 10 times, and 1 mL was taken as a sample.
[0045] (2) Preparation of template
[0046] The above samples were placed in a boiling water bath at 100°C for 10 minutes, in an ice bath for 5 minutes, and centrifuged at 12,000 r / min at 4°C for 5 minutes, and the supernatant was the DNA template.
[0047] (3) Loop-mediated isothermal amplification (LAMP) reaction:
[0048] Take 2 μL of the above template, 1 μL of each primer, 1 μL of Bst DNA polymerase (8U), 7.5 μL of amplification reaction solution, add water to a 25 μL reaction system, and react at a constant temperature of 63 °C for 1 h; then place it at 80 °C for 2 min to terminate the reaction.
[0049] The primers used are shown in SEQ ID NO: 1-4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com