Nucleic acid aptamer derivative and application thereof in preparation of medicament carrier
A technology of nucleic acid aptamers and derivatives, applied in biochemical equipment and methods, microbial measurement/testing, drug combination, etc., can solve problems such as difficult chemical modification and transformation, easy to cause immune response, and limit wide application, etc., to achieve Reduced toxicity, no immune activity and toxicity, high application value effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1. Preparation of nucleic acid aptamers and their derivatives
[0042] Three kinds of nucleic acid aptamer derivatives were artificially synthesized, and 1 GC, 29 GCs and 50 GCs were introduced into the 5' end of the nucleic acid aptamer TLS11a respectively. TLS11a-(GC) 1 As shown in sequence 3 of the sequence listing. TLS11a-(GC) 29 As shown in sequence 4 of the sequence listing. TLS11a-(GC) 50 As shown in sequence 5 of the sequence listing.
[0043] Three control nucleic acid aptamer derivatives were artificially synthesized, and 1 GC, 29 GC and 50 GC were introduced into the 5' end of the nucleic acid aptamer TD05, respectively. TD05-(GC) 1 As shown in sequence 6 of the sequence listing. TD05-(GC) 29 As shown in sequence 7 of the sequence listing. TD05-(GC) 50 As shown in sequence 8 of the sequence listing.
[0044] The nucleic acid aptamer TLS11a shown in sequence 1 of the sequence listing and the control nucleic acid aptamer TD05 shown in sequence 2 were...
Embodiment 2
[0053] 1. Aptamer TLS11a and aptamer derivative TLS11a-(GC) 29 preparation of
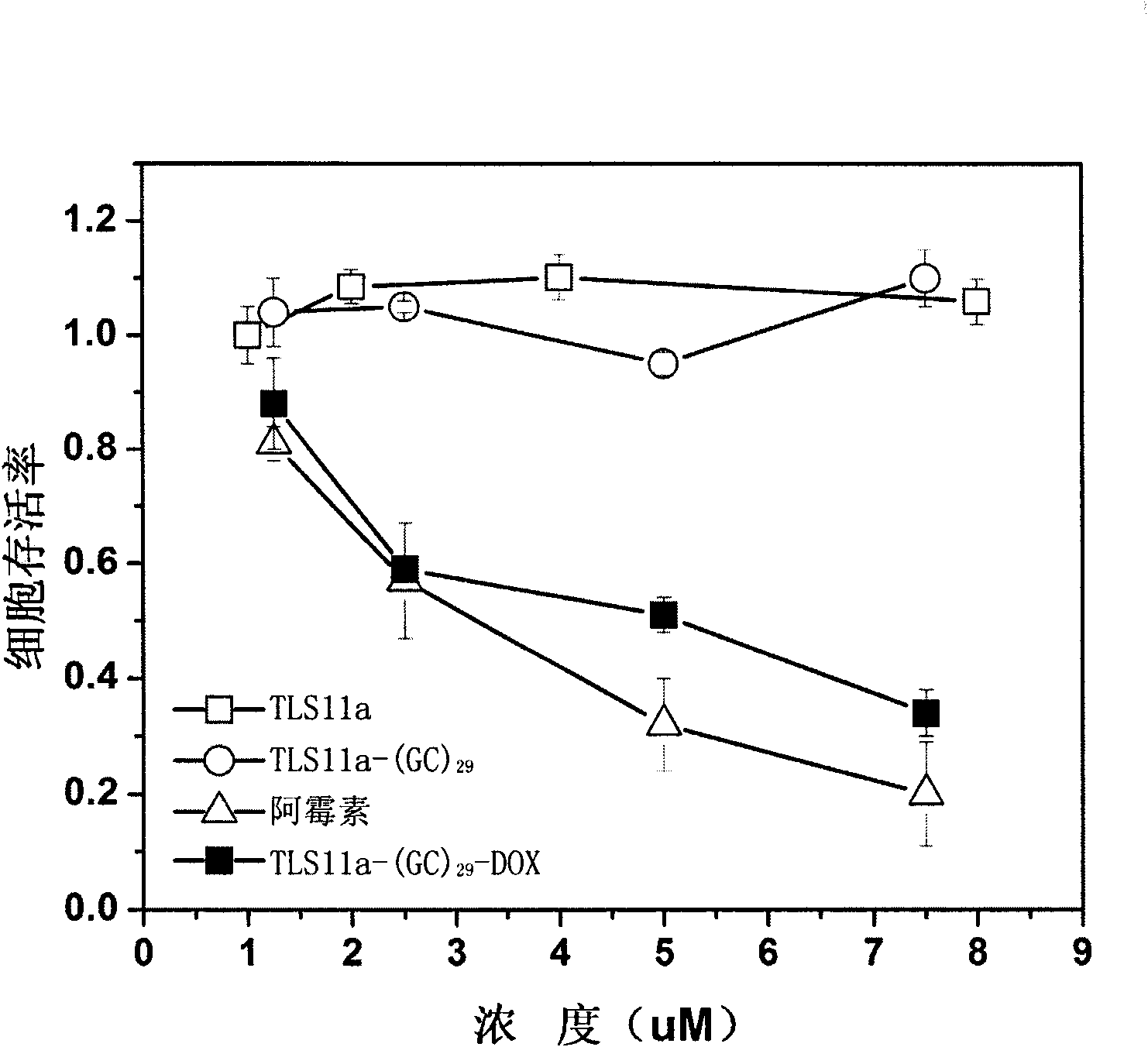
[0054] In order to embed more doxorubicin, a long GC sequence (composed of 29 GCs connected sequentially) was introduced at the 5' end of the nucleic acid aptamer TLS11a, thereby obtaining a nucleic acid aptamer derivative TLS11a-(GC ) 29 . TLS11a-(GC) 29 As shown in sequence 4 of the sequence listing.
[0055] The nucleic acid aptamer TLS11a shown in sequence 1 of the sequence listing was artificially synthesized. The artificially synthesized nucleic acid aptamer derivative TLS11a-(GC) shown in sequence 4 of the sequence listing 29 .
[0056] 2. Preparation and characterization of nucleic acid aptamer derivative-drug complex
[0057] 1. Preparation of nucleic acid aptamer derivative-drug complex
[0058] In the binding buffer, the nucleic acid aptamer derivative TLS11a-(GC) 29 Mix with doxorubicin at a ratio of 1:25 (molar ratio), and incubate with stirring at room temperature for 3 hours...
Embodiment 3
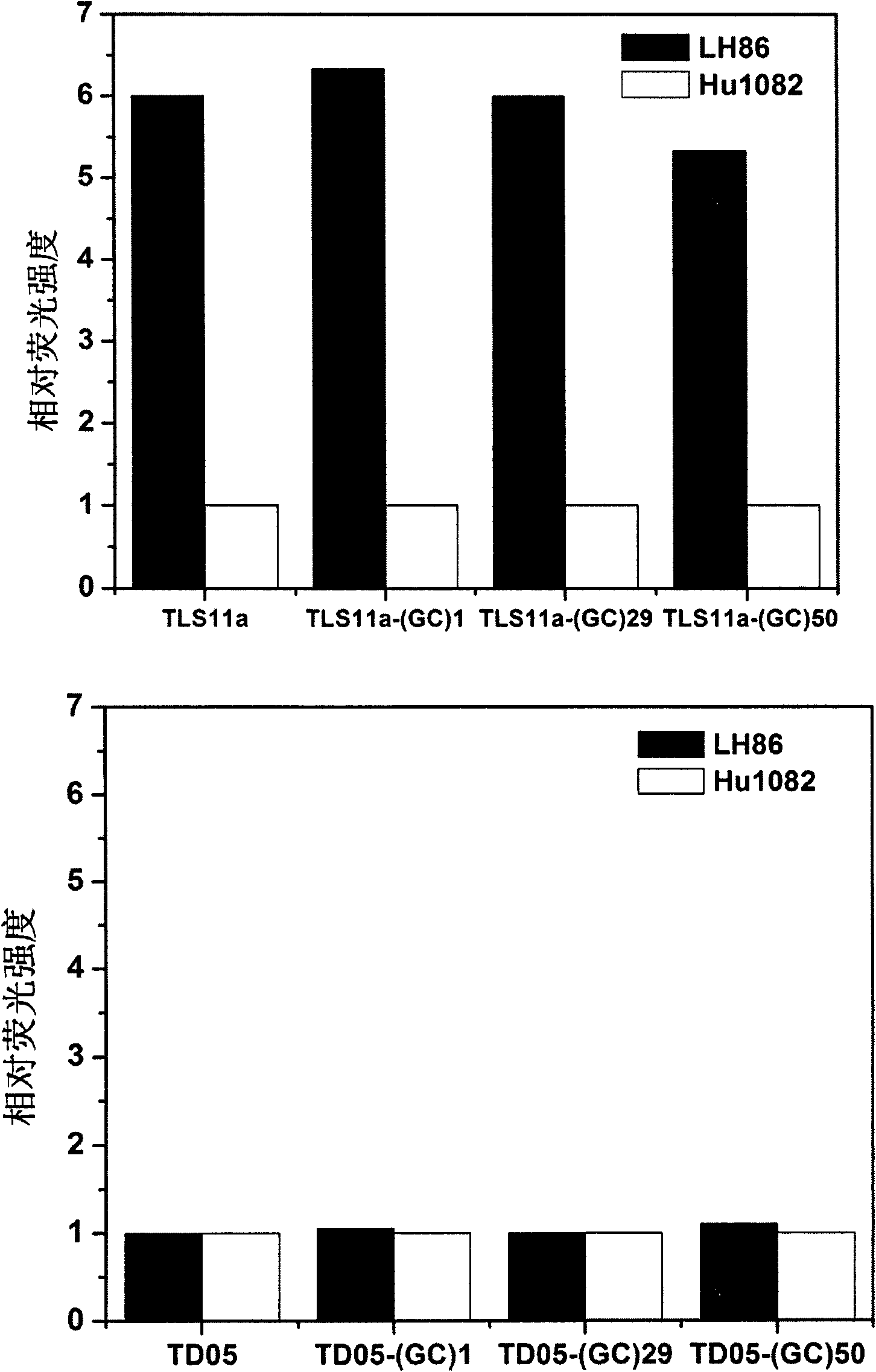
[0077] Determination of Chemosensitivity of Hepatoma Cell LH86 to Doxorubicin or Aptamer Derivative-Drug Complex Using CellTiter AQueous Single Solution Cell Proliferation Assay Kit (Promega, Madison, WI, USA) was performed. CellTiter AQueous single solution reagent: 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, internal salt; MTS and an electron coupling reagent ( thiophenazine, PES). The details are as follows: 100 μL of liver cancer cells LH86 (about 5×10 4 Cells / mL) were inoculated on 96-well plates (n=3) and grown overnight at 37°C (5% CO 2 , FBS-free DMEM medium), and then add 100 μL of the following DNA strands:
[0078] (1) TLS11a; set four concentration gradients, respectively with TLS11a-(GC) in the nucleic acid aptamer derivative-drug complex in (4) 29 The concentrations were the same, namely 40, 100, 200, 300 nM, respectively.
[0079] (2)TLS11a-(GC) 29 ; Four concentration gradients are set, respectively with TL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com