mRNA nucleoplasmic ratio variation-based method for identifying miRNA target gene and application thereof
An identification method and target gene technology, which can be used in biochemical equipment and methods, microbial determination/inspection, material inspection products, etc., and can solve problems such as miRNA discovery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
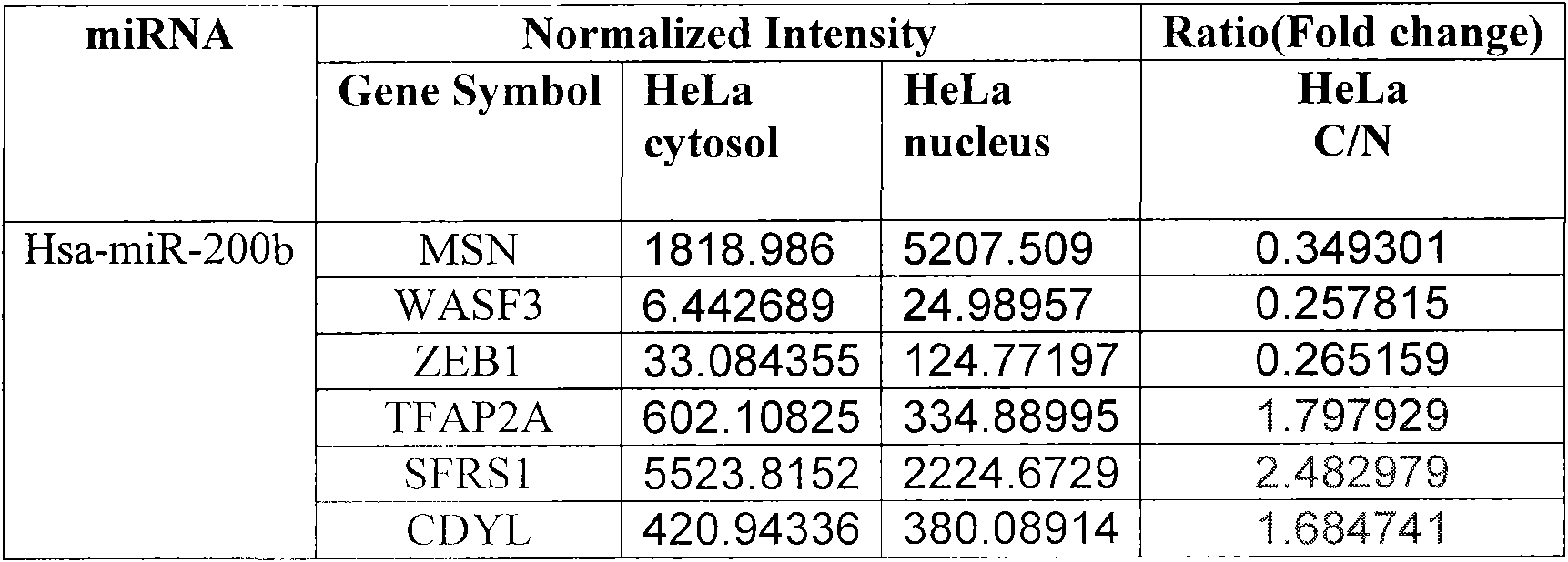
[0012] The following figure further describes the present invention by identifying the functional target gene of miR-200b in HeLa cells through specific application of the technical method of the present invention.
[0013] 1. Bioinformatics prediction:
[0014] Bioinformatics software such as TargetScan, miRanda, DIANAmicroT, PicTar, RNAHybrid, miRGen++, MiTarget, TarBase and other bioinformatics software were comprehensively used to predict the potential target genes of miR-200b, and the genes with the highest scores were randomly selected as targets ( figure 1 ).
[0015] 2. HeLa cytoplasmic and nuclear RNA extraction:
[0016] The cytoplasmic / nuclear RNA extraction kit (Norgen) was used to extract RNA from HeLa cells, and the main steps were as follows:
[0017] 1) Collect 3x10 6 Wash the cells once with ice-cold PBS, add 200 μl ice-cold lysate, and mix for 15 seconds.
[0018] 2) After the lysis is complete, centrifuge at room temperature at 16,000 rpm for 3 minutes, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com