Application of DNA target sequence of schistosoma japonicum retrotransposon in schistosomiasis diagnosis
A schistosome and DNA chip technology, applied in the fields of biotechnology and genetics, can solve the problem of limited retrotransposons of schistosomiasis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1 Obtaining retrotransposons and designing primers
[0062] 1. DNA extraction of Schistosoma japonicum adult samples
[0063] Using phenol extraction and ethanol precipitation of DNA:
[0064] 1) Take a single worm and put it into 200μl extraction buffer to wash the worm three times, and centrifuge at 2000g for 2min to separate the washing solution;
[0065] 2) Drain the washing solution, add 20 μl of extraction buffer, and use an RNAase-free sterilized small grinding stick to grind the worms until they are homogenized;
[0066] 3) Add extraction buffer to a final volume of 50 μl;
[0067] 4) Add 0.5 μl RNAase (10ug / μl), 5 μl SDS (10%), 0.5 μl protein digestion enzyme, and mix repeatedly with a gun;
[0068] 5) Digest in a compound water bath constant temperature oscillator at 110 rpm at 56°C for 1 hour;
[0069] 6) Add an equal volume of 50 μl phenol: chloroform: isopropanol 25:24:1 mixture, mix well, centrifuge at 10000 g for 5 min;
[0070] 7) Take the s...
Embodiment 2
[0098] Detection method and application based on retrotransposon SjCHGCS 19
[0099] Diagnosis remains central to schistosomiasis control activities. So far, etiological examination, that is, finding eggs in the feces of people in endemic areas, is still the only way and means to confirm the diagnosis of schistosomiasis. When certain pathogens infect the body, they also bring their genes into the human body. Theoretically speaking, as long as the pathogen exists, the body will have its nucleic acid substances. For the 25 retrotransposons in the Schistosoma japonicum genome listed in Table 1, the retrotransposon SjCHGCS 19 (SEQ ID NO: 19) was selected as a representative for in-depth research in this embodiment.
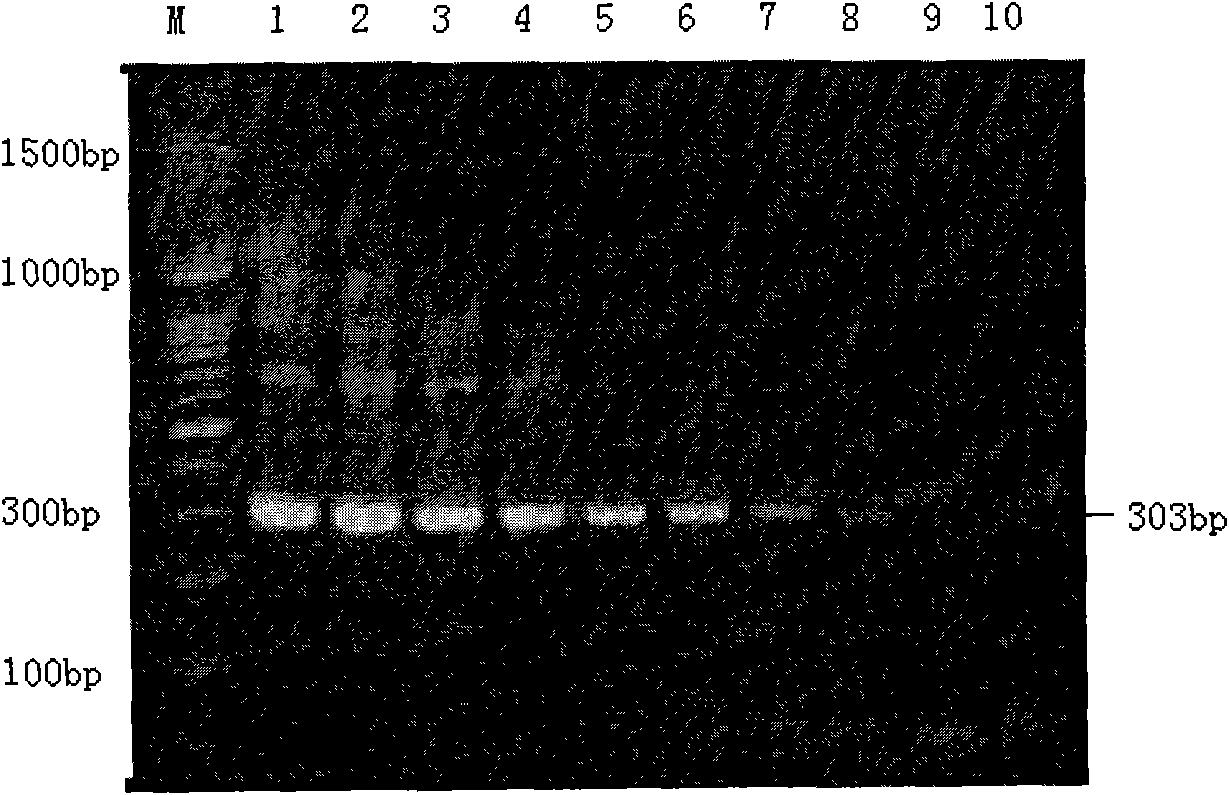
[0100] The results show that the retrotransposon SjCHGCS 19 polynucleotide is suitable as a detection marker for schistosomiasis, and is also suitable for preparing a kit or DNA chip for detecting schistosomiasis.
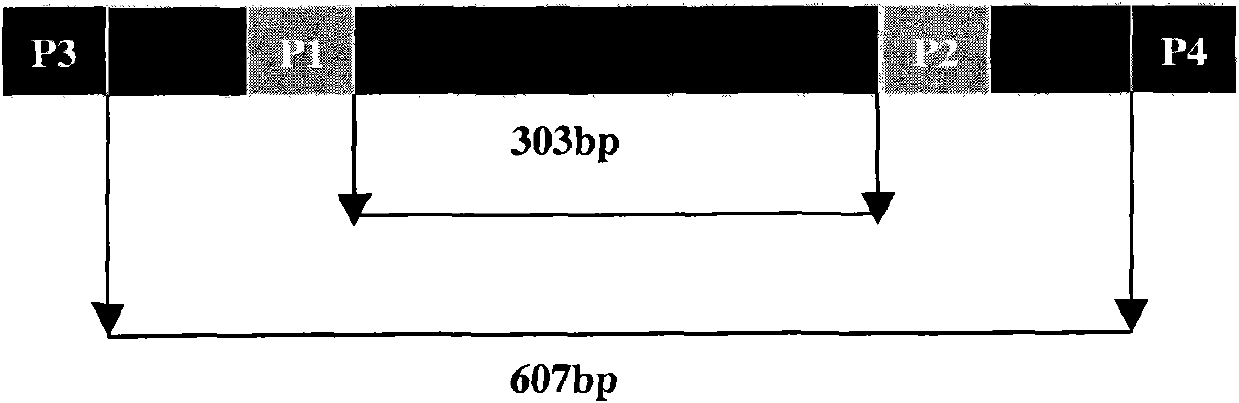
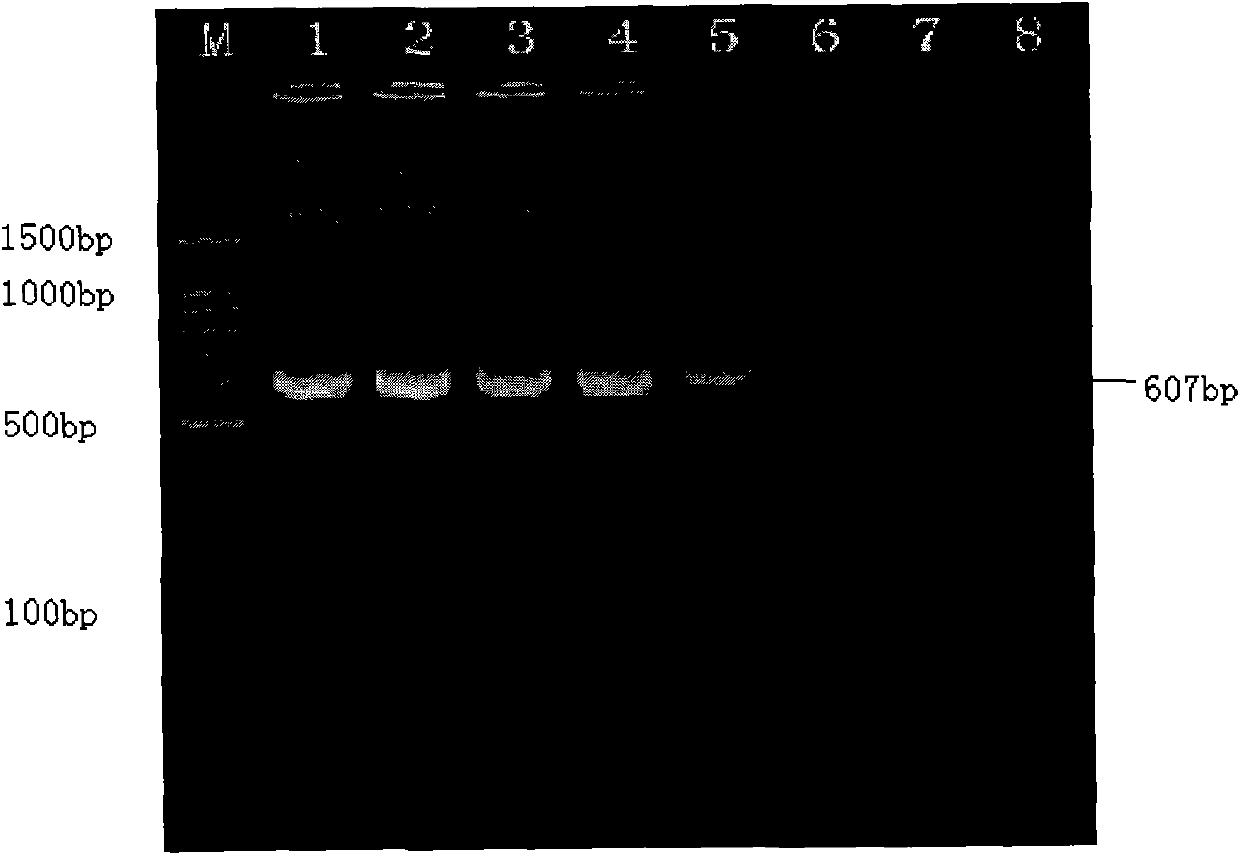
[0101] For example, the SjCHGCS 19P1-P2 fragment (...
Embodiment 3
[0110] Detection method and application based on retrotransposon SjCHGCS 10
[0111] Similar to Example 2, the retrotransposon SjCHGCS 10 (SEQ ID NO: 10) was selected as a representative for in-depth research in this example.
[0112] The detection sensitivity of the target sequence of Schistosoma japonicum retrotransposon SjCHGCS10.P1-P3 is as follows: Figure 8 As shown, the highest detection sensitivity is 1:10 6 (214.7fg / ul). The primers are SjCHGCS10.P1: GCCAGTGGGCATCTCCTT (SEQ ID NO: 30); SjCHGCS10.P3: CGGGCTGGCAATCAGTAA (SEQ ID NO: 31).
[0113] In addition, the SjCHGCS10.P1-P3 target sequence was used for DNA detection in serum of chronic patients with schistosomiasis japonicum. Similar to the detection results in Example 2, the positive detection rate is as high as 92% or more, and the reaction intensity of the amplified DNA band is also positively correlated with the degree of infection EPG.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com