Nucleic acid aptamer specifically combined with hepatitis C virus core protein and application thereof
A technology of hepatitis C virus and nucleic acid aptamer, which is applied in the determination/testing of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of harsh storage conditions, high cost, and high price, and achieve high application value , low cost, easy to store
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, preparation of related proteins and related solutions
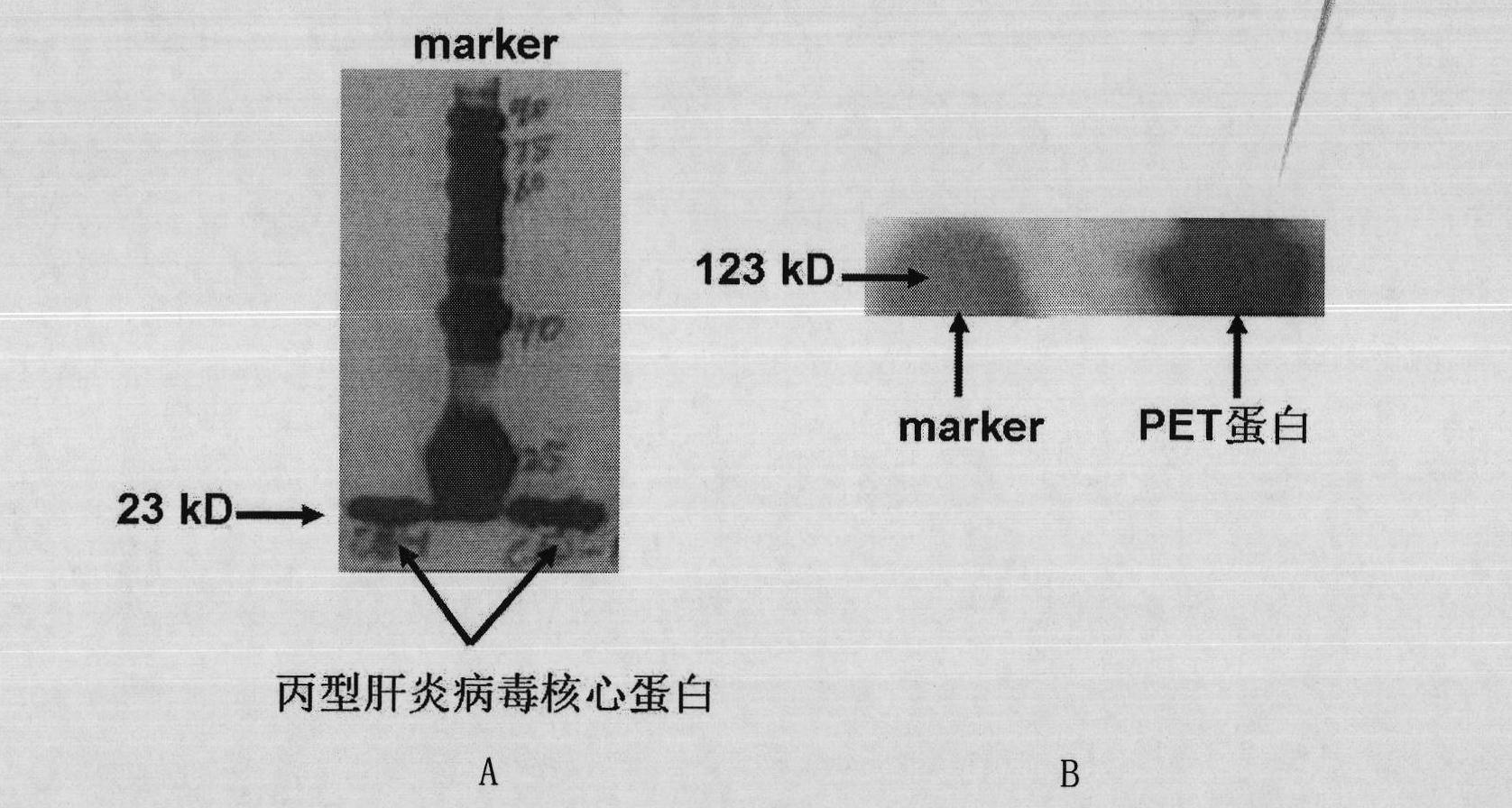
[0036] 1. Preparation of HCV core protein (target protein) with histidine tag
[0037] 1. Amplification of the coding gene of HCV core protein
[0038] Prepare the DNA shown in the sequence 4 of the sequence listing (the coding gene of the hepatitis C virus core protein, GENBANKACCESSION NO.HM566118.1, the hepatitis C virus core protein shown in the sequence 3 of the coding sequence listing), as PCR amplified As a template, a primer pair composed of primer 1 and primer 2 is used for PCR amplification to obtain a PCR amplification product.
[0039] Primer 1 (upstream primer): 5'-CGCGC GAATTC ATGAGCACGAATCCT-3';
[0040] Primer 2 (downstream primer): 5'-CTGCAG GGATCC AGAGGCCGGGACGGTCA-3';
[0041] EcoR1 restriction site and BamH1 restriction site were introduced into the 5' ends of the upstream and downstream primers respectively.
[0042] PCR amplification conditions: 95°C for 2min; 35 cycles of ...
Embodiment 2
[0071] Example 2, Screening and Preparation of Nucleic Aptamers
[0072] 1. Protein immobilization
[0073] 1. Take Ni-NTA agarose microbeads and place them in a 5ml centrifuge tube, remove the supernatant, and wash with PBS buffer three times;
[0074] 2. Disperse the microbeads in step 1 in the target protein (or control protein), incubate at room temperature for 1 hour, and centrifuge and wash with PBS buffer three times;
[0075] 3. Redisperse the microbeads from step 2 in 1ml of PBS buffer and store at 4°C for later use.
[0076] 2. Design of Random Nucleic Acid Library
[0077] A random nucleic acid library comprising 20 nucleotides at both ends and 40 nucleotides in the middle is designed as follows: 5'-ACGCTCGGATGCCACTACAG(N 40 ) CTCATGGACGTGCTGGTGAC-3'; N 40 Represents 40 random nucleotides.
[0078] 3. Screening of nucleic acid aptamers
[0079] 1. DNA library pretreatment
[0080] The random nucleic acid library is dissolved in binding buffer.
[0081] 2. An...
Embodiment 3
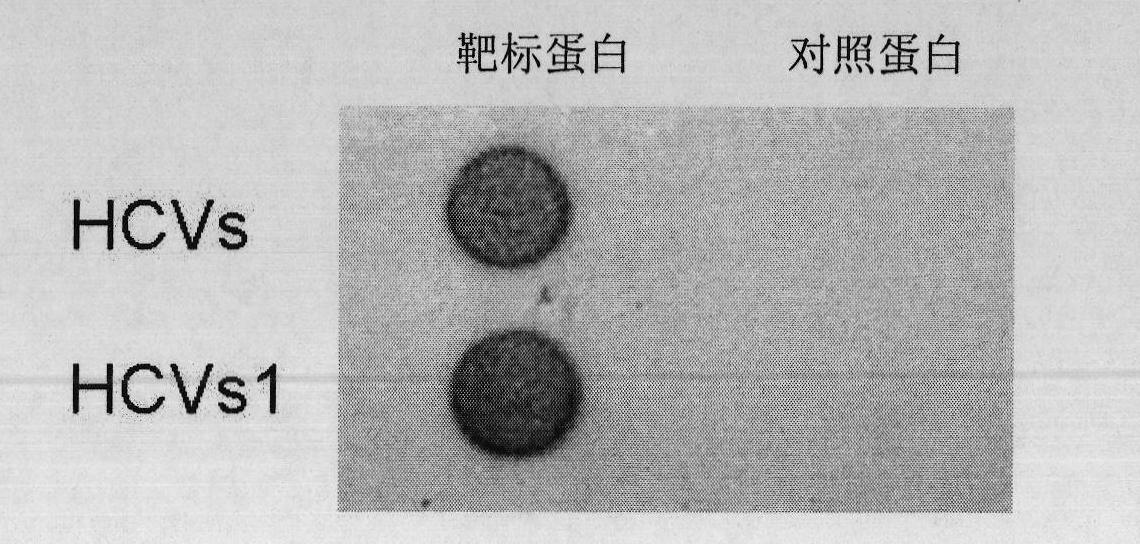
[0102] Embodiment 3, the specificity of nucleic acid aptamer (detected by fluorescein)
[0103] 1. FITC labeling of nucleic acid aptamers and random nucleic acid libraries
[0104] The aptamer HCVs prepared in Example 2 were labeled with fluorescein isothiocyanate (FITC).
[0105] The nucleic acid aptamer HCVs1 prepared in Example 2 was labeled with fluorescein isothiocyanate (FITC).
[0106] The random nucleic acid library prepared in Example 2 was labeled with fluorescein isothiocyanate (FITC).
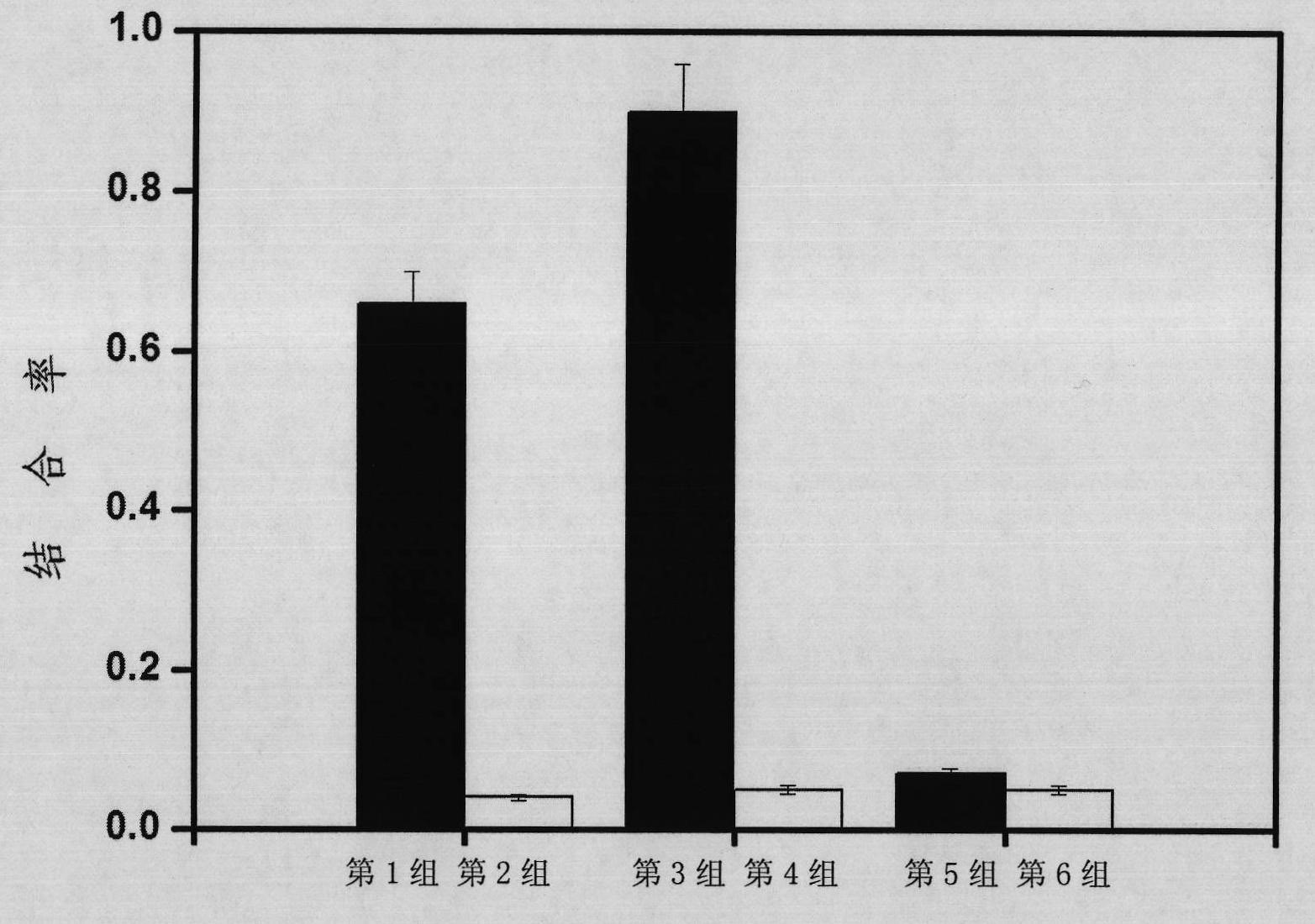
[0107] 2. The specificity of nucleic acid aptamers
[0108] 1. Protein immobilization
[0109] (1) Immobilization of target protein
[0110] ①Put 200ul Ni-NTA agarose microbeads in a 5ml centrifuge tube, remove the supernatant, and wash with PBS buffer three times;
[0111] ② Disperse the microbeads in step ① in 1 mL of the target protein, incubate at room temperature for 1 h, and centrifuge and wash with PBS buffer three times;
[0112] ③ Re-disperse the microbeads from step ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com