Molecular marker for single fruit weight main-effect quantitative trait loci (QTL) of Dangshansu pear fruit and application thereof
A Dangshansu pear and molecular marker technology, which is applied in the field of molecular genetics and breeding, can solve the problems of positioning and molecular markers are not developed, and achieve good application value, improve accuracy and efficiency, and improve the effect of accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
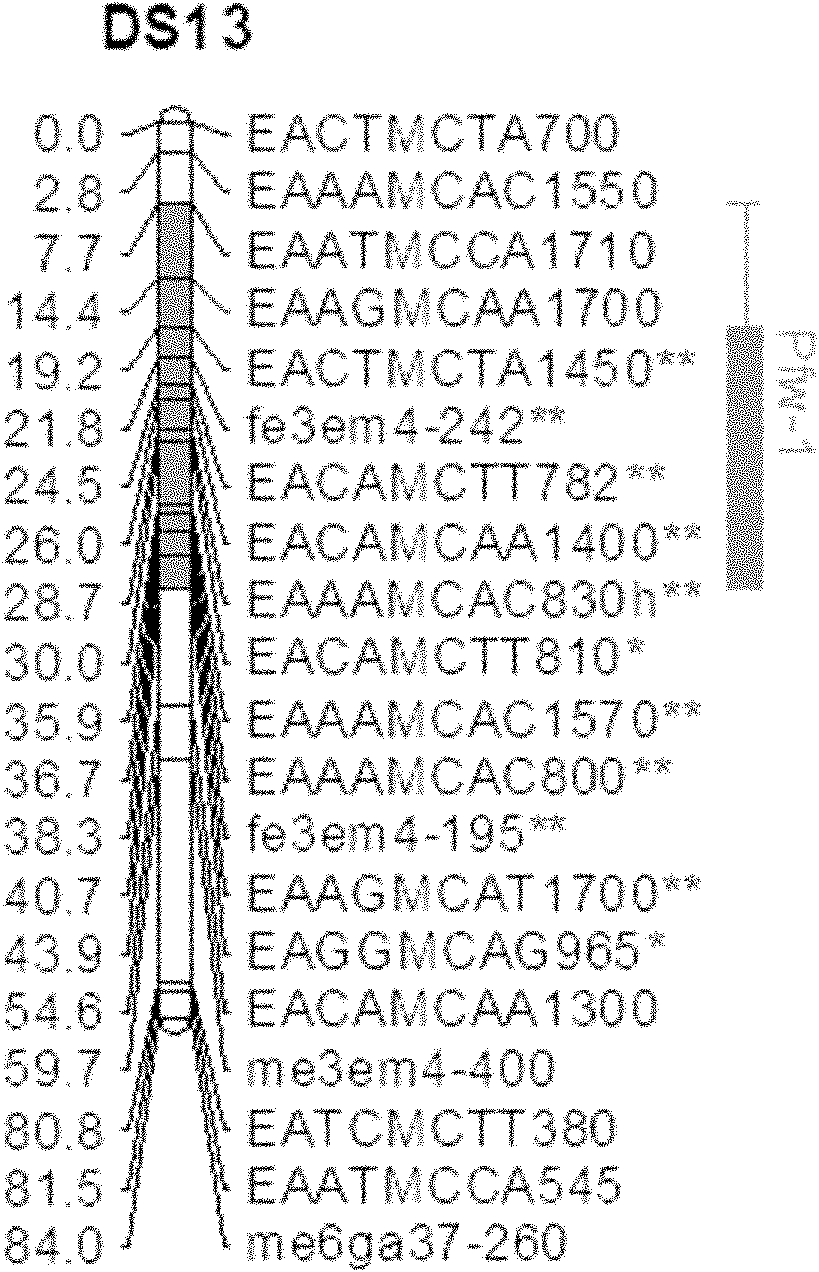
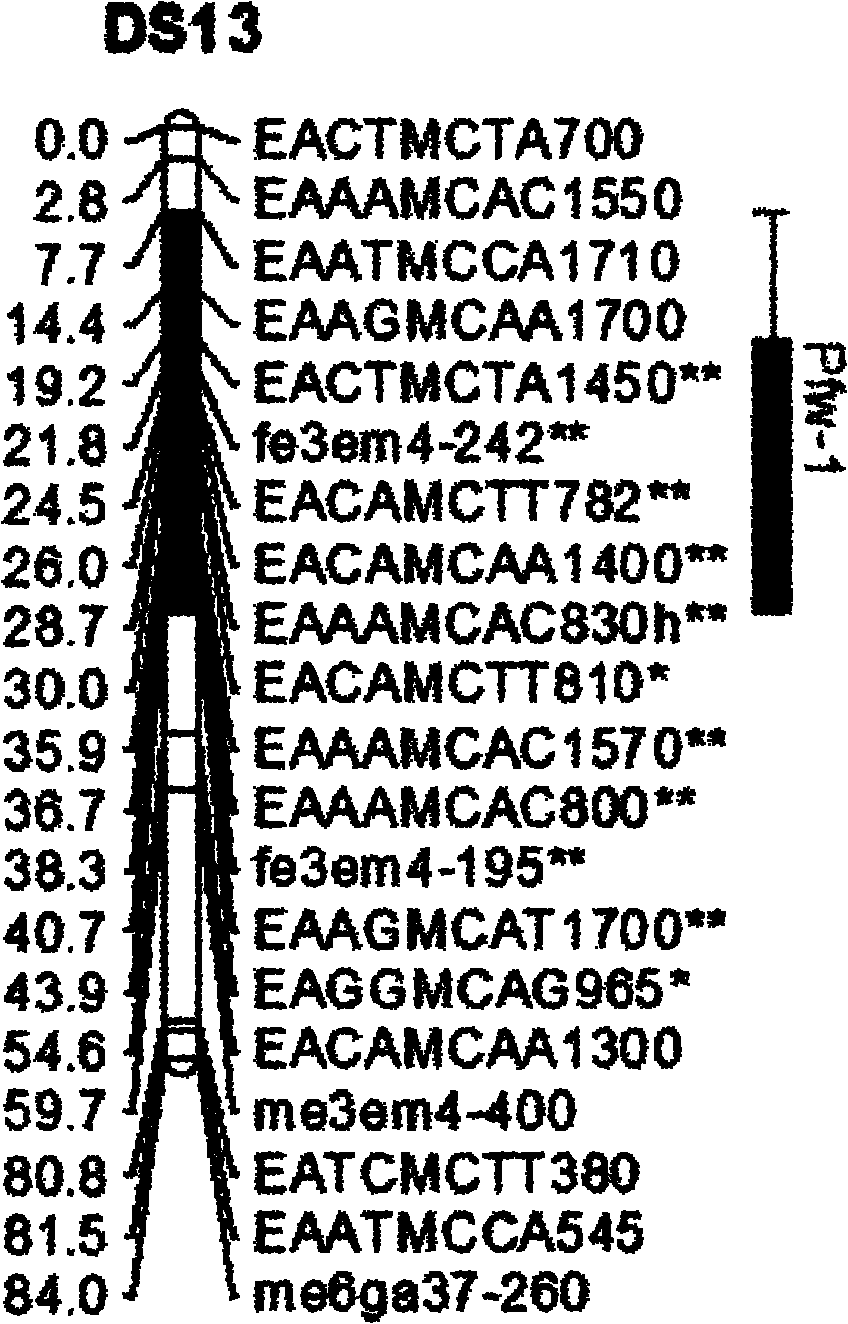
[0030] a) Using the hybrid combination of two pear varieties 'August Hong' and 'Dangshansu pear' in my country, 97 F 1 group.
[0031] b) Analyze the two parents and their F 1Population, statistical analysis of genetic types of polymorphic sites between parents in offspring populations. A total of 732 polymorphic marker loci, including 23 SSRs, S gene loci, 226 SRAPs, and 482 AFLPs, were finally obtained, and then linkage analysis was performed on them using Jionmap3.0 mapping software to construct a table containing Genetic linkage map of 'Dangshansuli' with 240 polymorphic markers.
[0032] Using SSR, SRAP, and AFLP molecular marker methods, the 'August Red' pear and 'Dangshansu pear' and their offspring were analyzed, and the genetic types of the polymorphic sites between the parents were statistically analyzed in the offspring population.
[0033] For the experimental procedure of SSR labeling, refer to the method of Yomamoto T. et al. (Euphytica, 2002, 124:129-137); fo...
Embodiment 2
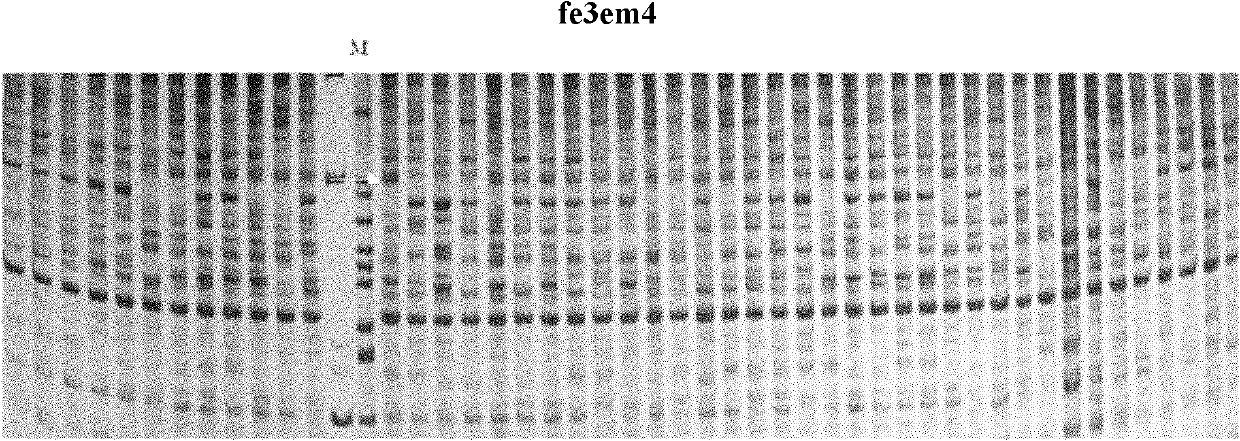
[0045] Using the screened molecular markers linked to the main effect QTL for single fruit weight of ‘Dangshansu pear’, the 97 strains of ‘August Hong’ and ‘Dangshansu pear’ 1 Preliminary testing was carried out on the hybrid progenies to test the practical value of this method in molecular marker-assisted selection of pear fruit single fruit weight. Use the genomic DNA of the 97 offspring and SRAP primers fe3 and em4 to carry out PCR reactions, and the PCR amplification system for SRAP analysis is a total volume of 20ul: which contains 0.2mmol L -1 dNTPs, 0.3 μmol L -1 Upstream and downstream primers, 2.0mmol L -1 MgCl 2 , 1 unit of rTaq enzyme, 50ng of DNA template, 1×Buffer. The PCR reaction program was: 94°C for 3min; 35 cycles: 94°C for 40s, 55°C for 50s, 72°C for 1min; 72°C for 10min, 10°C for 10min. The results of PCR amplification were electrophoresed on 8% non-denaturing polyacrylamide gel (results such as figure 2 ), after electrophoresis, determine whether the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com