Escherichia coli engineering bacteria for expressing recombinant sucrose phosphorylase
A sucrose phosphorylase, Escherichia coli technology, applied in recombinant DNA technology, enzymes, bacteria and other directions, can solve the problems of complex process, high cost, low extraction rate, etc., and achieve high purification efficiency, low cost and high specificity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
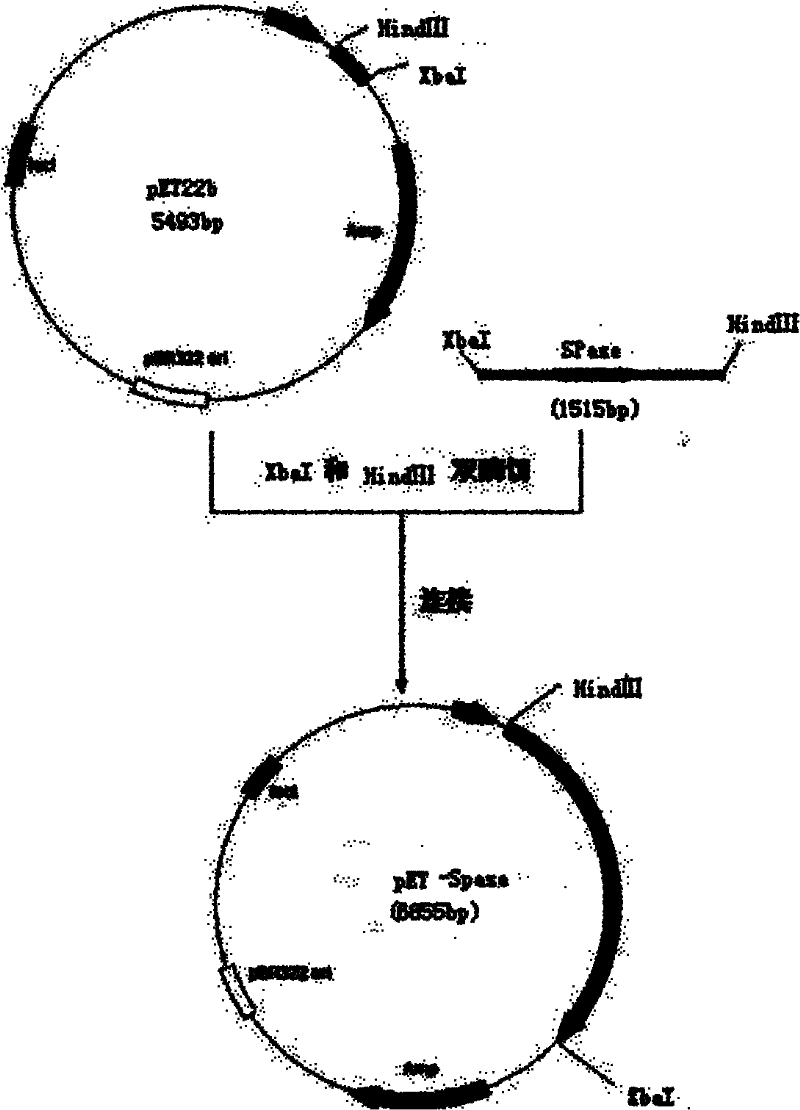
[0050] Example 1 Construction of Escherichia coli sp1515 genetic engineering bacteria.
[0051] 1. Primer design
[0052] According to the SPase gene sequence (GenBank Accession NO.D90314.1) of Leuconostoc mesenteroides ATCC12291, a pair of primers for amplifying the SPase gene were designed with Primerpremier 5, namely the upstream primer P1 and the downstream primer P2:
[0053] Upstream primer P1: 5'-GC TCTAGA CGCAATCGTTACAGTTATTACTAGC-3';
[0054] Downstream primer P2: 5'-CCC AAGCTT GTTCTGAGTCAAATTATCACTGCTC-3'.
[0055] Xba I and Hind III restriction enzyme sites (underlined) were introduced at the 5' ends of P1 and P2, respectively, and protective bases were added.
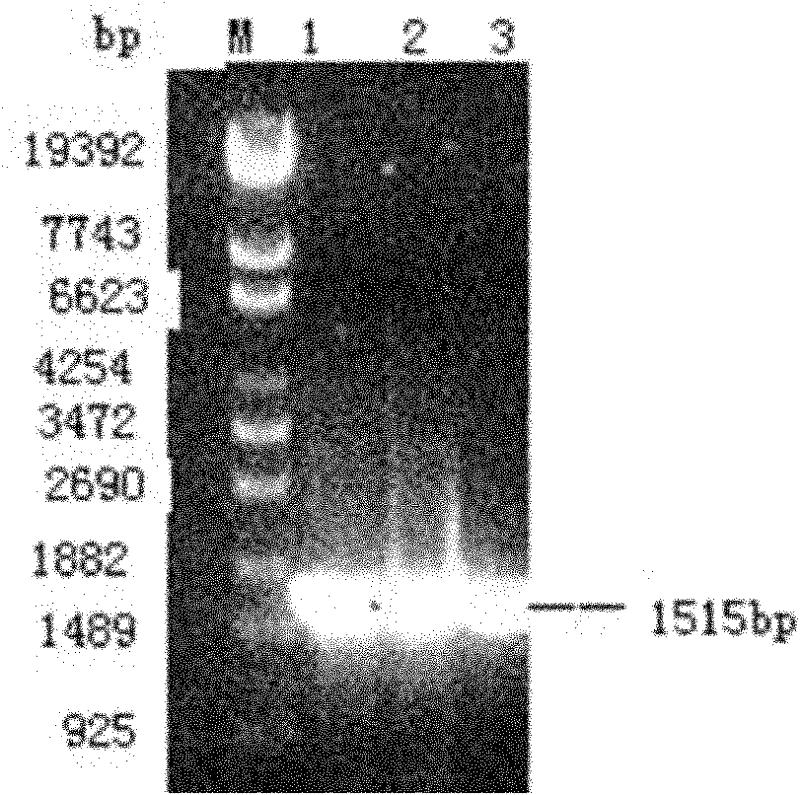
[0056] 2. PCR amplification of Leuconostoc enteromenis SPase fragment
[0057] (1) Genomic DNA extraction of Leuconostoc enterolis
[0058] a) Streak and activate Leuconostoc mesenteroides ATCC 12291 stored in glycerol cryopreservation tubes, inoculate a single colony in 5ml MRS medium and culture ov...
Embodiment 2
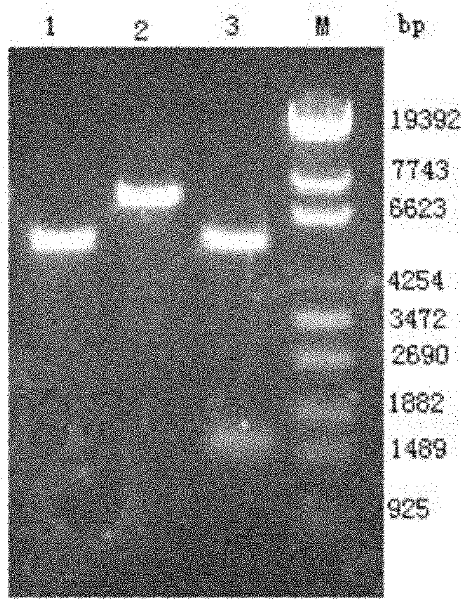
[0093] Example 2 Expression of Escherichia coli sp1515 genetic engineering bacteria
[0094] 1. IPTG induces the expression of Escherichia coli sp1515 genetically engineered bacteria
[0095] (1) Pick a single colony of the recombinant bacteria and culture it overnight in LB medium, and use the bacterial sample containing the empty vector and E.coli BL21(DE3) as controls;
[0096] (2) The next day, the experimental group and the control group were inoculated into fresh 50mL LB medium according to the volume ratio of 3% inoculum, and cultured at 37°C with shaking at 200r / min;
[0097] (3) Wait until the bacterial solution grows to OD 600 At 0.6-0.8, add the inducer IPTG to a final concentration of 0.5mmol / L, and induce at 30°C for 6 hours;
[0098] (4) The bacteria liquid was centrifuged (4° C., 8000 r / min, 15 min) to collect the bacteria.
[0099] 2. SDS-PAGE electrophoresis detection of expression products
[0100] (1) Wash the expression product collected by the above ce...
Embodiment 3
[0109] The influence of embodiment 3 different induction temperature, different IPTG concentration and different induction time on SPase recombinant protein expression
[0110] The expression conditions of genetically engineered bacteria were optimized through further research on induction temperature, IPTG induction concentration and induction time.
[0111] 1. The influence of different induction temperature
[0112] (1) Pick a single colony of recombinant bacteria and culture it overnight in LB medium;
[0113] (2) The next day, the seed solution was inoculated into fresh 50mL LB medium according to the volume ratio of 3% inoculum, and cultured with shaking at 37°C and 200r / min;
[0114] (3) Wait until the bacterial solution grows to OD 600 At 0.6-0.8, IPTG was added to a final concentration of 0.1 mmol, and then induced at 20°C, 25°C, 30°C, and 35°C for 10 hours. The result is as Figure 5 shown.
[0115] 2. Effect of different IPTG concentrations
[0116] (1) (2) ar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com